Search Count: 59
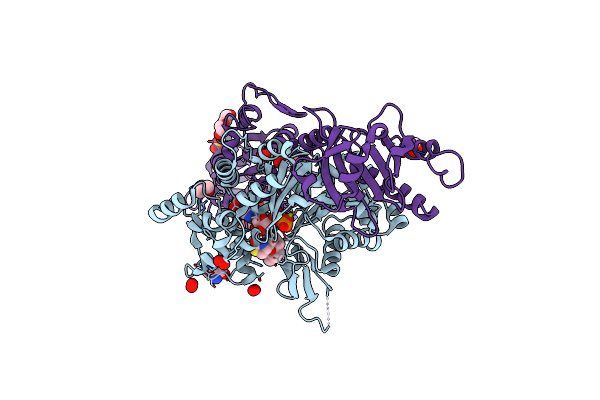 |
Chimera Of The Inactive Exoy Nucleotidyl Cyclase Domain From Vibrio Nigripulchritudo Martx Toxin, With The Double Mutation K3528M And K3535I, Fused To A Proline-Rich-Domain (Prd) And Profilin, Bound To Latrunculin B-Adp-Mg-Actin
Organism: Vibrio nigripulchritudo, Homo sapiens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2023-09-20 Classification: TOXIN Ligands: ADP, MG, LAB, TRS, GOL, PEO, SO4, PEG |
 |
Chimera Of Exoy Nucleotidyl Cyclase Domain From Vibrio Nigripulchritudo Fused To A Proline-Rich-Domain (Prd) And Profilin, Bound To Adp-Mg-Actin And A Sulfate Ion
Organism: Vibrio nigripulchritudo, Homo sapiens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-09-20 Classification: TOXIN Ligands: ADP, MG, TRS, GOL, PEG, PEO, SO4 |
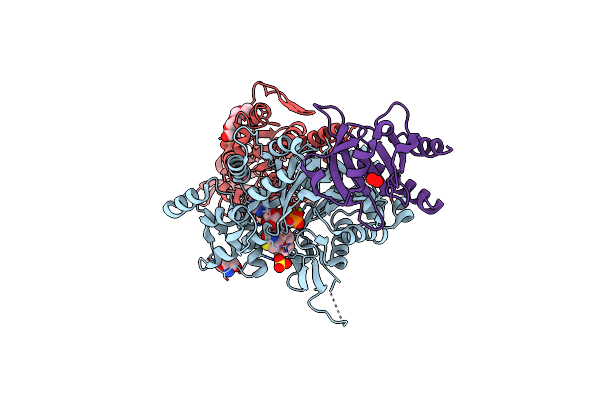 |
Exoy Nucleotidyl Cyclase Domain From Vibrio Nigripulchritudo Martx Toxin, Bound To Atp-Mg-Actin, Human Profilin 1 And A Sulfate Ion
Organism: Homo sapiens, Vibrio nigripulchritudo, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-09-20 Classification: TOXIN Ligands: ATP, MG, LAB, TRS, SO4, PEO, PG4, GOL |
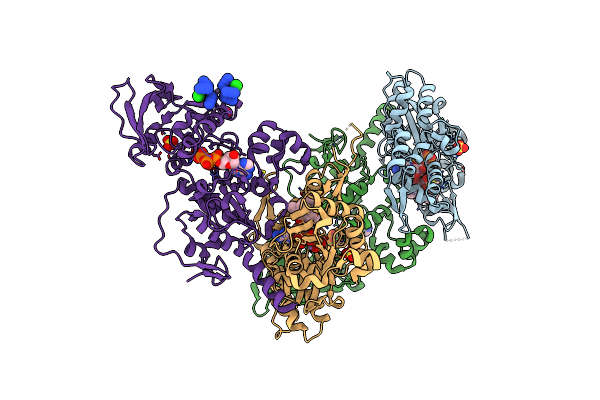 |
Exoy Nucleotidyl Cyclase Domain From Vibrio Nigripulchritudo Martx Toxin, Bound To Latrunculin-B-Atp-Mg-Actin, And 3'-Deoxyadenosine-5'-Triphosphate And 2 Mg Ions
Organism: Vibrio nigripulchritudo, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-09-20 Classification: TOXIN Ligands: LAB, ATP, MG, SO4, 3AT, MN, AZI, CL |
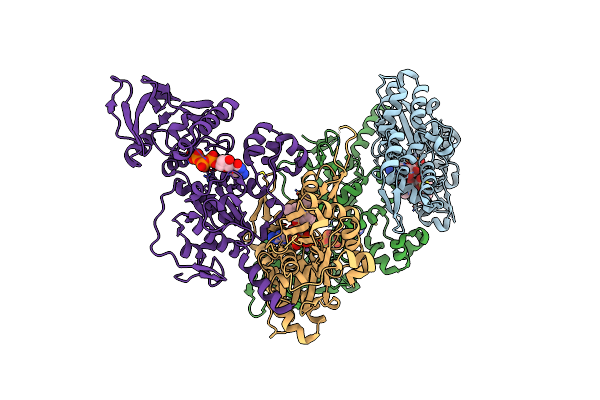 |
Exoy Nucleotidyl Cyclase Domain From Vibrio Nigripulchritudo Martx Toxin (Residue Q3455 To L3863) In Complex With 3'Deoxyctp And Two Manganese Cations Bound To Latrunculin-B-Adp-Mn-Actin
Organism: Vibrio nigripulchritudo sfn135, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-09-20 Classification: TOXIN Ligands: LAB, ADP, MN, CH1 |
 |
Exoy Nucleotidyl Cyclase Domain From Vibrio Nigripulchritudo Martx Toxin, Bound To Latrunculin-B-Atp-Mg-Actin, And 3'-Deoxyadenosine-5'-Triphosphate And 2 Mg Ions
Organism: Vibrio nigripulchritudo, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-09-20 Classification: TOXIN Ligands: LAB, ATP, MG, 3AT, SO4, GOL, PEO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: METAL BINDING PROTEIN Ligands: CL, MG |
 |
Cryo-Em Structure Of A Prefusion Stabilized Sars-Cov-2 Spike (D614N, R682S, R685G, A892P, A942P And V987P)(S-Closed Trimer)
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of A Prefusion Stabilized Sars-Cov-2 Spike (D614N, R682S, R685G, A892P, A942P And V987P)(One Up Trimer)
Organism: Severe acute respiratory syndrome coronavirus 2, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-09-25 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
Organism: Homo sapiens, Human spumaretrovirus, Pyrobaculum filamentous virus 1
Method: ELECTRON MICROSCOPY Release Date: 2019-09-25 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-05-17 Classification: CELL CYCLE Ligands: SO4, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-05-17 Classification: CELL CYCLE Ligands: SO4, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-17 Classification: CELL CYCLE Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2017-05-17 Classification: CELL CYCLE |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-01-21 Classification: HYDROLASE/DNA Ligands: ZN |
 |
Organism: Bungarus fasciatus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-11-26 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: EDO, NAG |
 |
Crystal Structure Of The C-Terminal Region Of Yeast Ctf4, Selenomethionine Protein.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-04-30 Classification: DNA REPLICATION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-04-30 Classification: DNA REPLICATION |
 |
Crystal Structure Of The Carboxy-Terminal Domain Of Yeast Ctf4 Bound To Pol Alpha.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-04-30 Classification: DNA REPLICATION |

