Search Count: 149
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: METAL BINDING PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: OT5 |
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Streptomyces collinus
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Streptomyces collinus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: METAL BINDING PROTEIN Ligands: AKG, FE |
 |
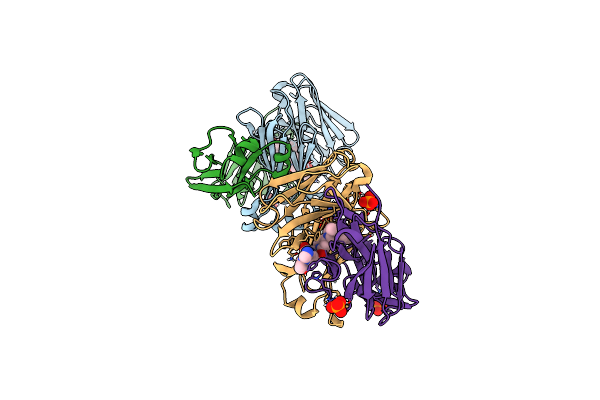
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 1-(1-(Imidazo[1,5-A]Pyrazin-8-Yl)Azetidin-3-Yl)-3-(2-(6-Methyl-4-(3-Methyl-3-Phenylpyrrolidin-1-Yl)-2-Oxopyridin-1(2H)-Yl)Ethyl)Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AHP |
 |
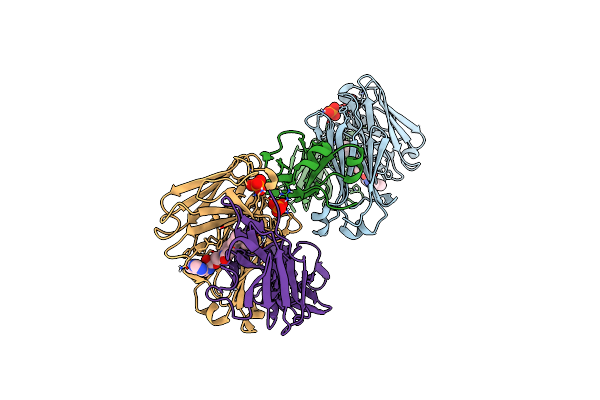
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 3-(2-(4-(3-Ethyl-3-Phenylpyrrolidin-1-Yl)-6-Methyl-2-Oxopyridin-1(2H)-Yl)Ethyl)-8-(Imidazo[1,5-A]Pyrazin-8-Yl)-1,3,8-Triazaspiro[4.5]Decan-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AH0 |
 |
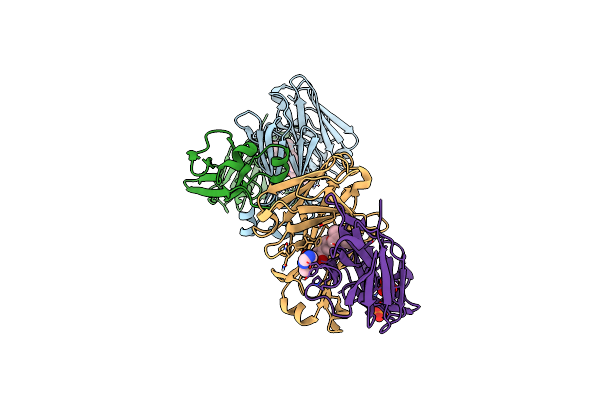
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 2-(6-Methyl-2-Oxo-4-(2-Phenylpropoxy)Pyridin-1(2H)-Yl)Ethyl (3-(1H-Imidazol-4-Yl)Benzyl)Carbamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AH2 |
 |
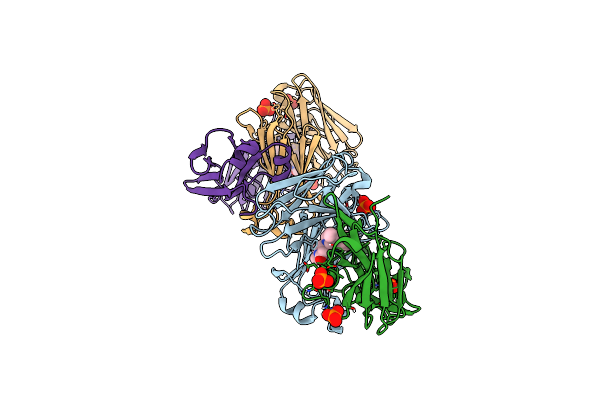
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 2-(1-Methyl-3-Oxo-5-(2-Phenylpropoxy)Isoindolin-2-Yl)Ethyl (3-(1H-Imidazol-4-Yl)Benzyl)Carbamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: A1AH6, PO4 |
 |
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 4-(3-Ethyl-3-Phenylpyrrolidin-1-Yl)-1,6-Dimethylpyridin-2(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AH1 |
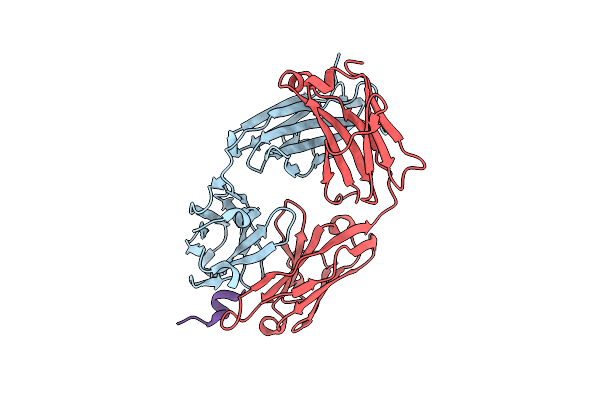 |
The Co-Crystal Structure Of The Fab Fragment Of Ab-1080 With Nav1.7 Vsdii Peptide
Organism: Oryctolagus cuniculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
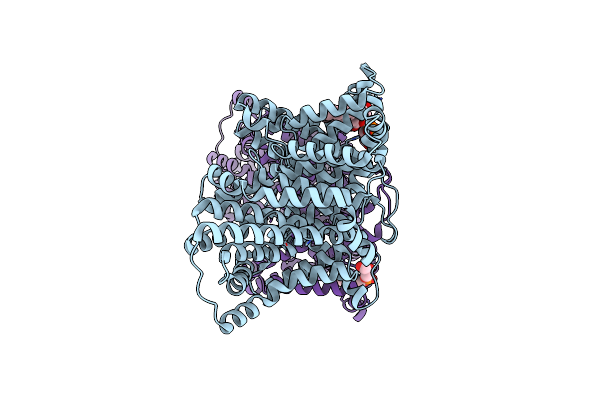 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Parallel Dimer)
Organism: Haemophilus influenzae rd kw20
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: PGT, NA |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
Organism: Haemophilus influenzae rd kw20
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN |

