Search Count: 15
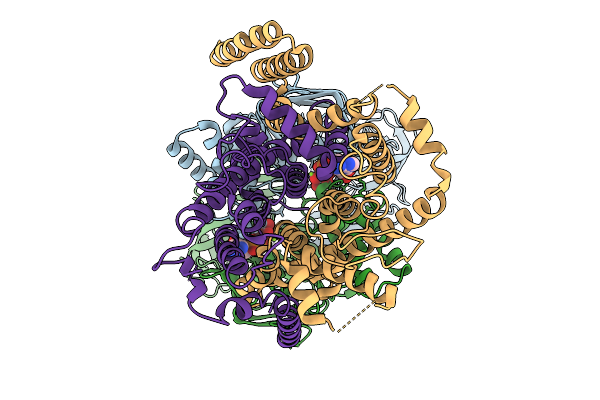 |
Cryo-Em Structure Of A Bacterial Prototype Atp-Binding Cassette Transporter Malfgk2.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN Ligands: MG, ADP, VO4 |
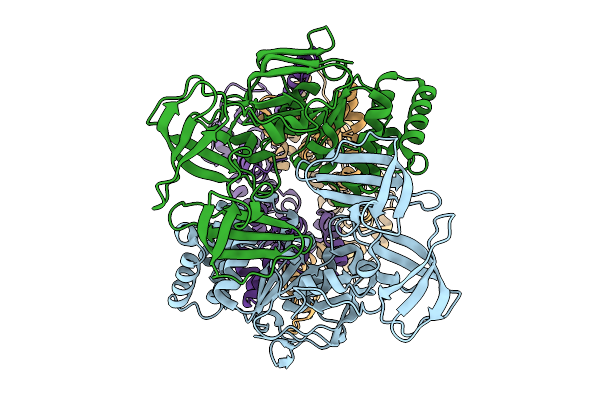 |
Cryo-Em Structure Of A Bacterial Prototype Atp-Binding Cassette Transporter Malfgk2.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN |
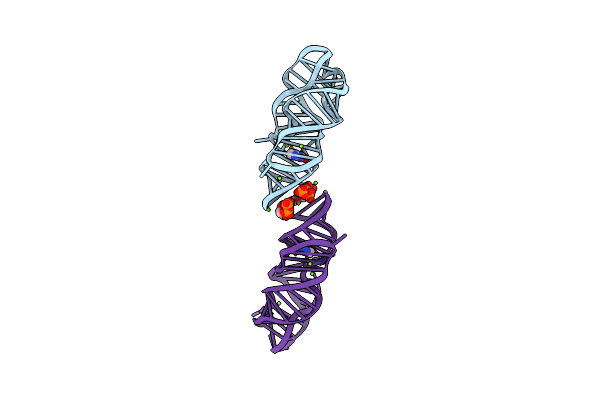 |
Crystal Structure Of P. Beijingensis Xanthine-Ii Riboswitch In Complex With Xanthine
Organism: Paenibacillus beijingensis
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: RNA Ligands: GTP, MG, XAN |
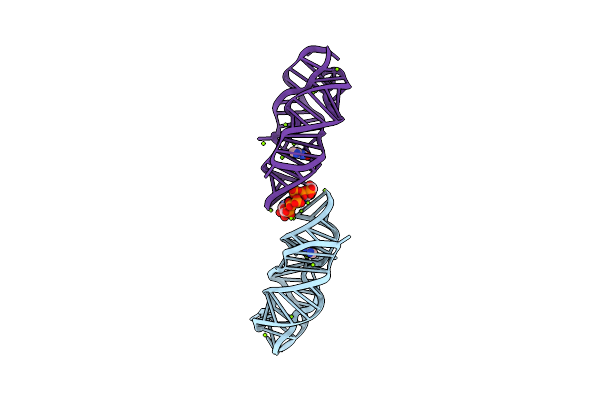 |
Organism: Paenibacillus beijingensis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-06-04 Classification: RNA Ligands: GTP, MG, XAN |
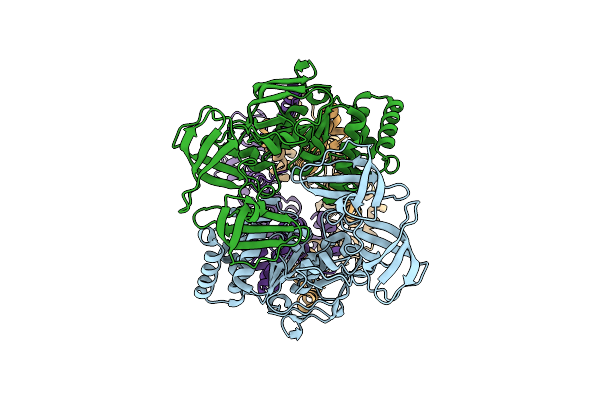 |
Cryo-Em Structure Of A Bacterial Prototype Atp-Binding Cassette Transporter Malfgk2.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: TRANSPORT PROTEIN |
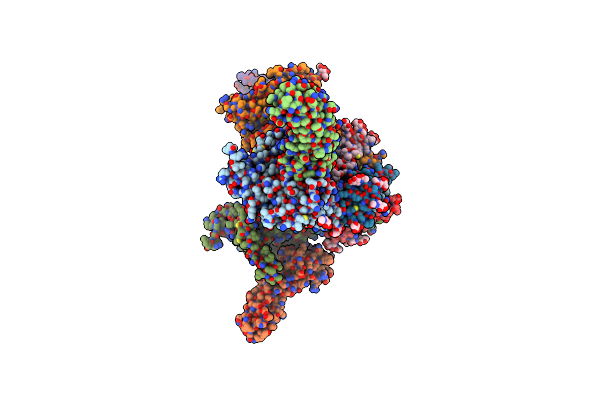 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRUS Ligands: NAG |
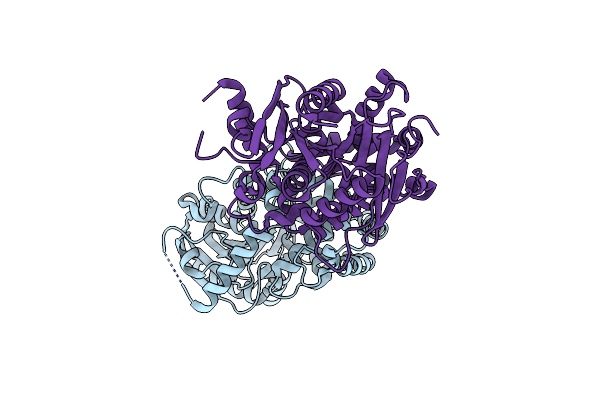 |
Crystal Structure Of An Epoxide Hydrolase Mutant A250Ic/L344V From Aspergillus Usamii E001 At 2.17 Angstroms Resolution
Organism: Aspergillus usamii
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-02-07 Classification: HYDROLASE |
 |
The Dna Gyrase B Atp Binding Domain Of Pseudomonas Aeruginosa In Complex With Compound 12X
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-02 Classification: ISOMERASE Ligands: DMS, SO4, NA, EZ6 |
 |
The Dna Gyrase B Atp Binding Domain Of Pseudomonas Aeruginosa In Complex With Compound 12O
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-09-02 Classification: ISOMERASE Ligands: EZ9, SO4, CL |
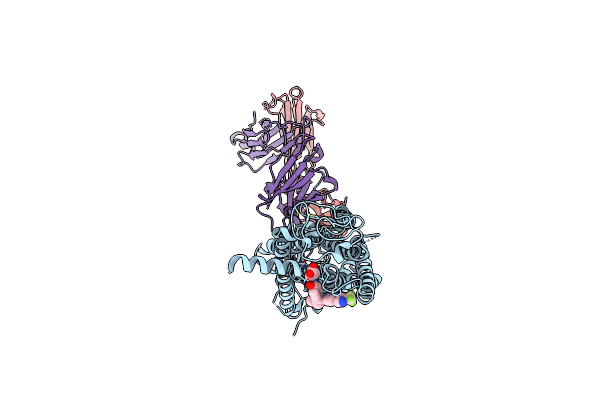 |
Crystal Structure Of Full Length Human Glp1 Receptor In Complex With Fab Fragment (Fab7F38)
Organism: Homo sapiens, Clostridium pasteurianum, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-03-18 Classification: MEMBRANE PROTEIN Ligands: NAG, ZN, 97Y |
 |
Solution Structure Of The Hydrophobin Mpg1 From The Rice Blast Fungus Magnaporthe Oryzae
Organism: Magnaporthe oryzae 70-15
Method: SOLUTION NMR Release Date: 2016-05-18 Classification: STRUCTURAL PROTEIN |
 |
An Nmr/Saxs Structure Of The Pki Domain Of The Honeybee Dicistrovirus, Israeli Acute Paralysis Virus (Iapv) Ires
|
 |
Organism: Neurospora crassa
Method: SOLUTION NMR Release Date: 2014-08-06 Classification: STRUCTURAL PROTEIN |
 |
Organism: Neurospora crassa
Method: SOLUTION NMR Release Date: 2013-06-05 Classification: STRUCTURAL PROTEIN |
 |
Identification Of The Key Regions That Drive Functional Amyloid Formation By The Fungal Hydrophobin Eas
Organism: Neurospora crassa
Method: SOLUTION NMR Release Date: 2012-01-25 Classification: STRUCTURAL PROTEIN |

