Search Count: 7
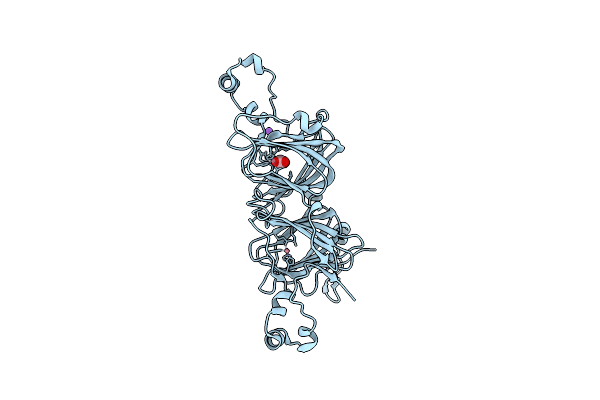 |
The Substrate Binding Mode And Chemical Basis Of A Reaction Specificity Switch In Oxalate Decarboxylase
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-04-06 Classification: LYASE Ligands: CO, OXL, NA |
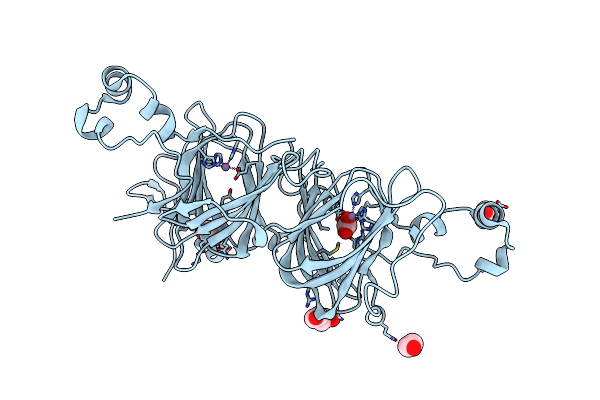 |
A Structural Element That Modulates Proton-Coupled Electron Transfer In Oxalate Decarboxylase
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2012-04-25 Classification: LYASE Ligands: MN, CO3, EDO, PEG |
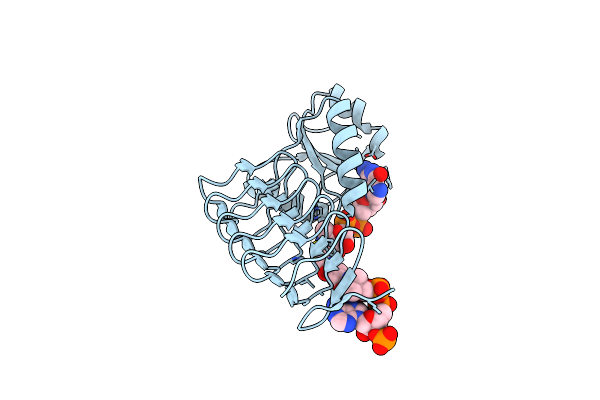 |
X-Ray Crystal Structure Of Perb From Caulobacter Crescentus In Complex With Coa And Gdp-Perosamine At 1.0 Angstrom Resolution
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2012-04-04 Classification: TRANSFERASE Ligands: COA, JB2, CL |
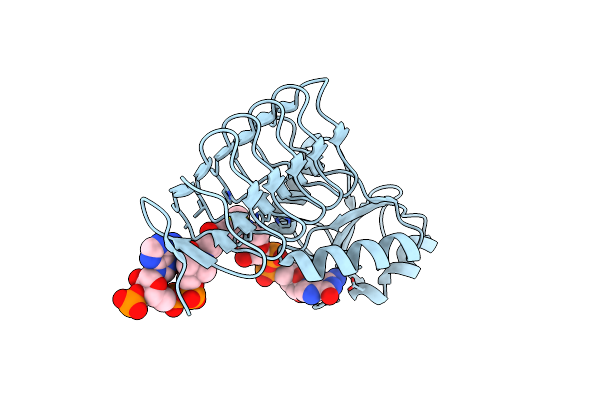 |
X-Ray Crystal Structure Of Perb From Caulobacter Crescentus In Complex With Coenzyme A And Gdp-N-Acetylperosamine At 1 Angstrom Resolution
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2012-04-04 Classification: TRANSFERASE Ligands: COA, JB3, CL |
 |
X-Ray Structure Of Gdp-Perosamine N-Acetyltransferase In Complex With Transition State Analog At 0.9 Angstrom Resolution
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2012-04-04 Classification: TRANSFERASE Ligands: JBT, CL |
 |
X-Ray Crystal Structure Of The H141N Mutant Of Perosamine N-Acetyltransferase From Caulobacter Crescentus In Complex With Coa And Gdp-Perosamine
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-04-04 Classification: TRANSFERASE Ligands: JB2, COA, NA, CL |
 |
X-Ray Crystal Structure Of The H141A Mutant Of Gdp-Perosamine N-Acetyl Transferase From Caulobacter Crescentus In Complex With Coa And Gdp-Perosamine
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2012-04-04 Classification: TRANSFERASE Ligands: JB2, COA, NA, CL |

