Search Count: 27
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MOTOR PROTEIN Ligands: ATP, ADP |
 |
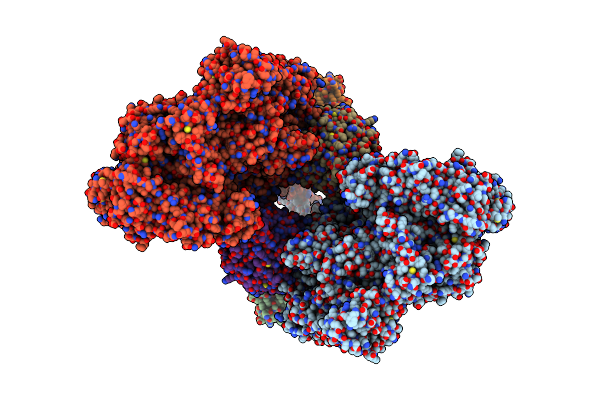
Symmetry Expansion Of Yeast Cytoplasmic Dynein-1 Bound To Lis1 In The Chi Conformation.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: MOTOR PROTEIN Ligands: ATP, ADP |
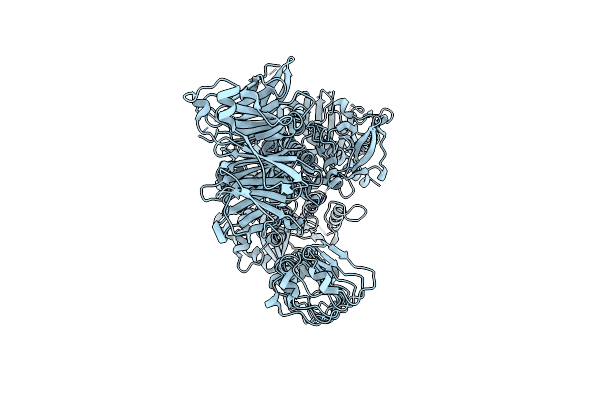 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: GDP |
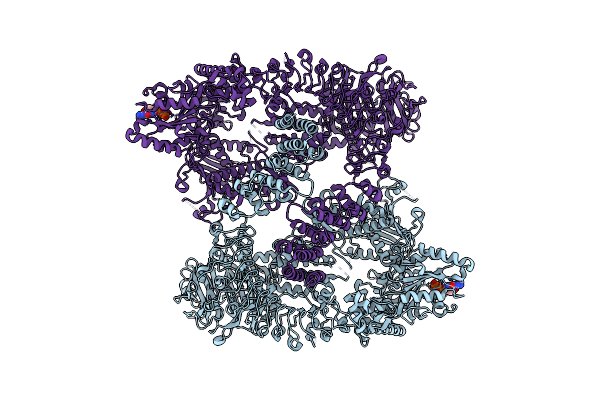 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: GDP |
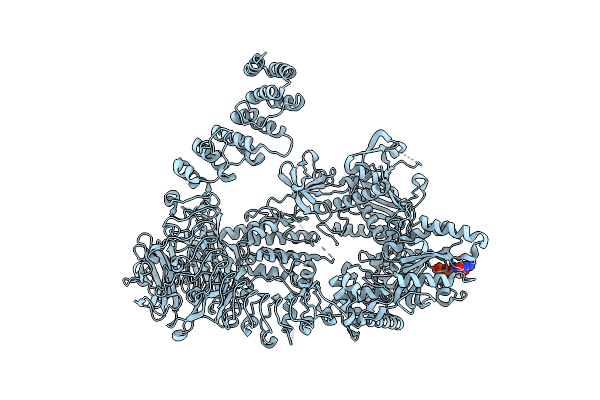 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSFERASE Ligands: GDP |
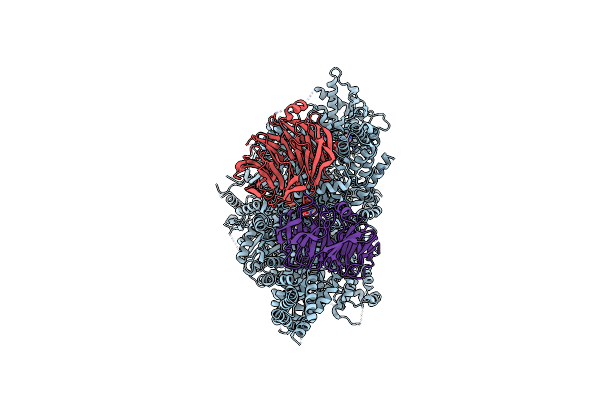 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: MOTOR PROTEIN Ligands: ADP, ATP |
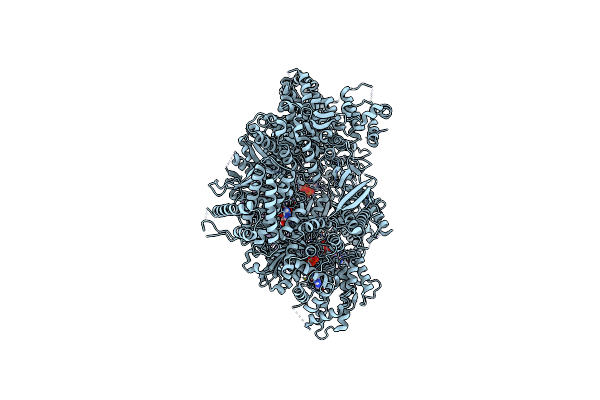 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: MOTOR PROTEIN Ligands: ADP, ATP |
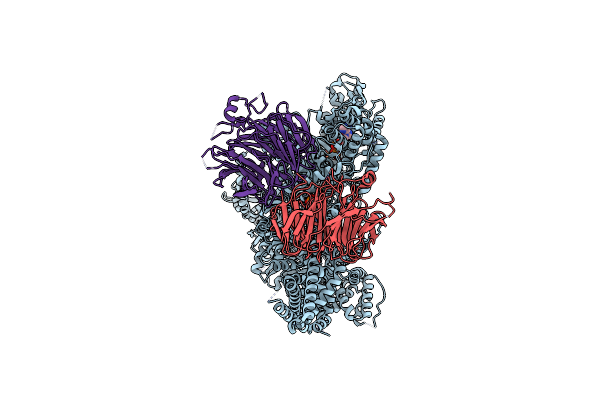 |
Structure Of Yeast Cytoplasmic Dynein With Aaa3 Walker B Mutation Bound To Lis1
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: MOTOR PROTEIN Ligands: ATP, ADP, MG |
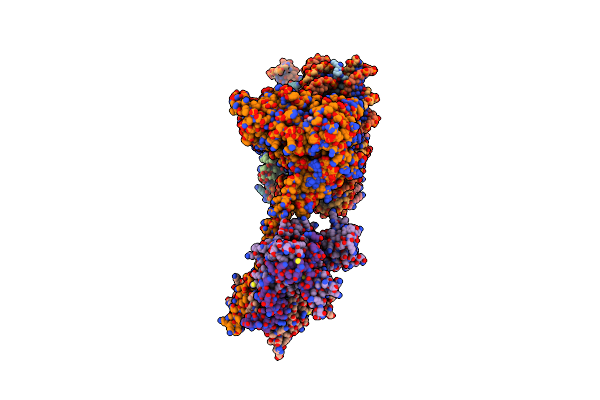 |
Cryo-Em Structure Of Sth1-Arp7-Arp9-Rtt102 Bound To The Nucleosome In Adp Beryllium Fluoride State
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: MOTOR PROTEIN Ligands: MG, ADP, BEF, ATP |
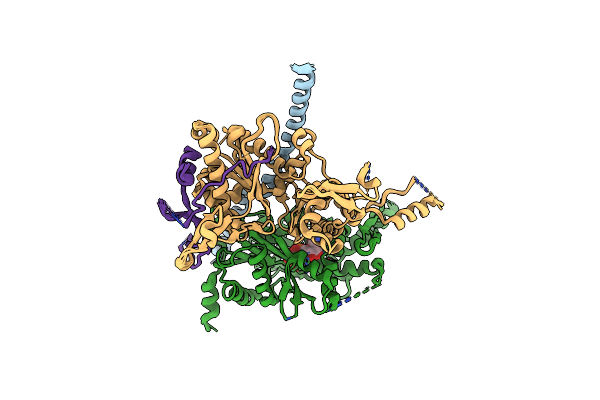 |
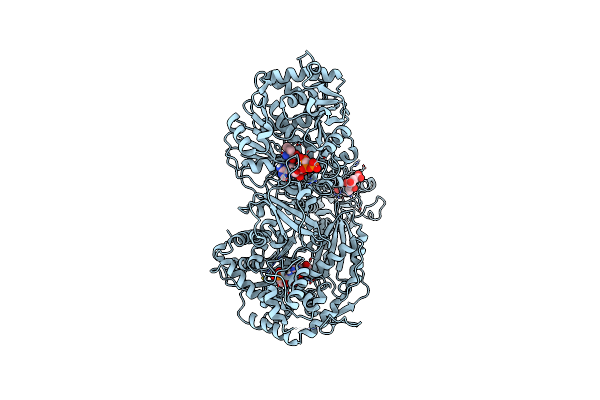
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: MOTOR PROTEIN Ligands: ATP |
 |
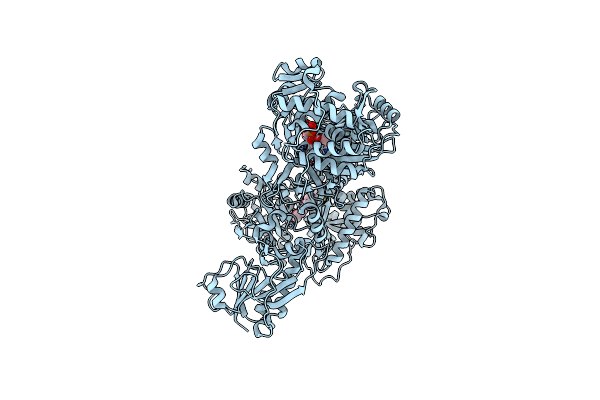
Crystal Structure Of A 4-Domain Construct Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: JQG, FON, APC, VAL, PO4, MG |
 |
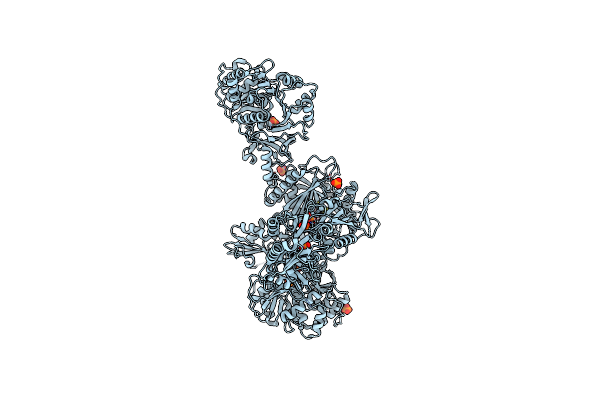
Crystal Structure Of A 4-Domain Construct Of A Mutant Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: 9EF, APC, VAL |
 |
Crystal Structure Of A 5-Domain Construct Of Lgra In The Substrate Donation State
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: PNS, PO4 |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:6.00 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: PNS |
 |
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:6.00 Å Release Date: 2019-11-20 Classification: LIGASE Ligands: DG9 |
 |
Crystal Structure Of The Formyltransferase Psej From Anoxybacillus Kamchatkensis
Organism: Anoxybacillus kamchatkensis g10
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-10-17 Classification: TRANSFERASE Ligands: MES, SO4 |
 |
Crystal Structure Of The Formyltransferase Psej From Anoxybacillus Kamchatkensis Soaked With Udp-4-Amino-4,6-Dideoxy-L-Altnac
Organism: Anoxybacillus kamchatkensis g10
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2018-10-17 Classification: TRANSFERASE Ligands: F5P |
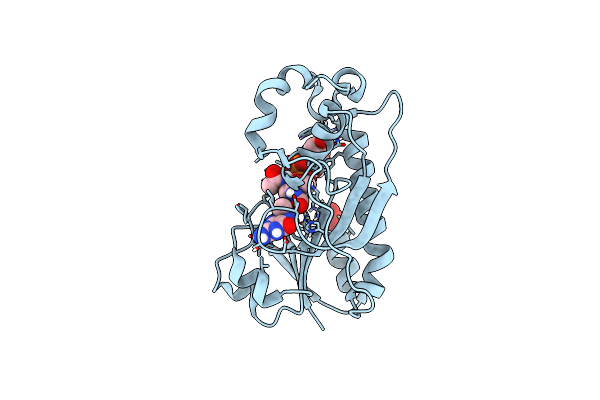 |
Crystal Structure Of The Formyltransferase Psej From Anoxybacillus Kamchatkensis In Complex With Udp-4,6-Dideoxy-4-Formamido-L-Altnac And Tetrahydrofolate
Organism: Anoxybacillus kamchatkensis g10
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-10-17 Classification: TRANSFERASE Ligands: SO4, F5G, 1YJ |
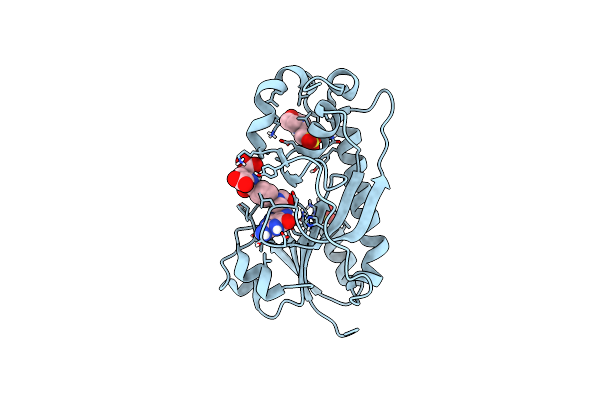 |
Crystal Structure Of The Formyltransferase Psej From Anoxybacillus Kamchatkensis With N10-Formyltetrahydrofolate
Organism: Anoxybacillus kamchatkensis g10
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-17 Classification: TRANSFERASE Ligands: MES, SO4, 1YA |
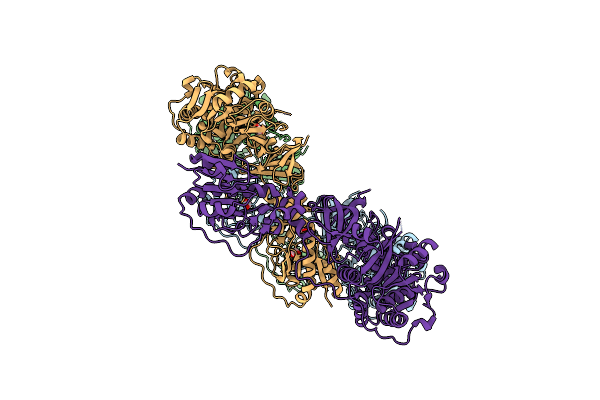 |
Crystal Structure Of The Lgra Initiation Module Excluding The Asub Domain: F-A-Delta-Sub
Organism: Brevibacillus parabrevis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2016-10-12 Classification: LIGASE Ligands: FMT |

