Search Count: 17
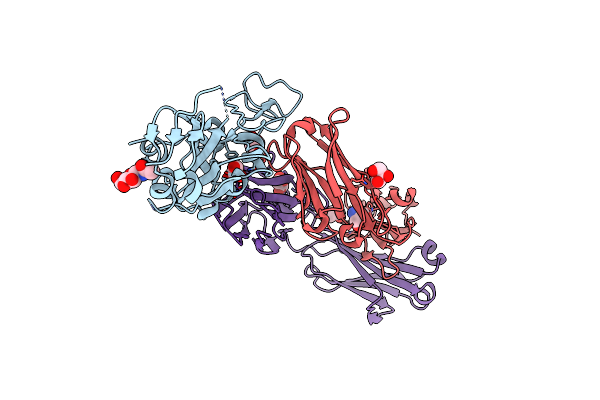 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain (Rbd) In Complex With 1D1 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-17 Classification: VIRAL PROTEIN Ligands: NAG, GOL, TRS |
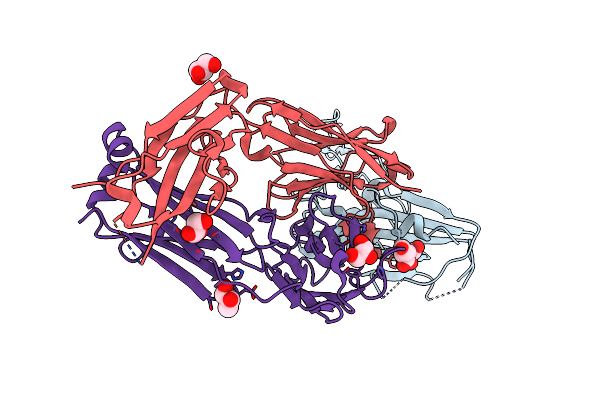 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain (Rbd-Beta Variant) In Complex With 3D2 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-17 Classification: VIRAL PROTEIN Ligands: NAG, GOL |
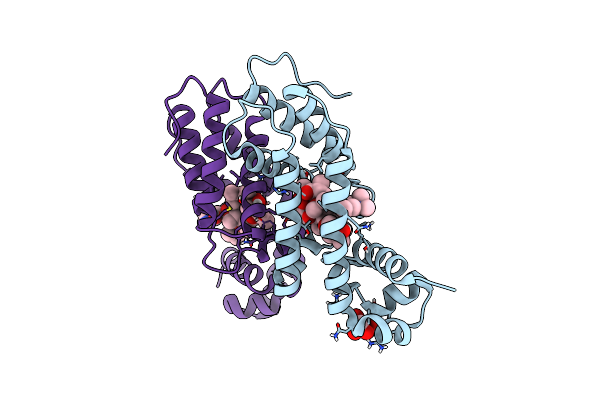 |
Directed Evolution Of A Biosensor Selective For The Macrolide Antibiotic Clarithromycin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-19 Classification: DNA BINDING PROTEIN/ANTIBIOTIC Ligands: CTY, FLC |
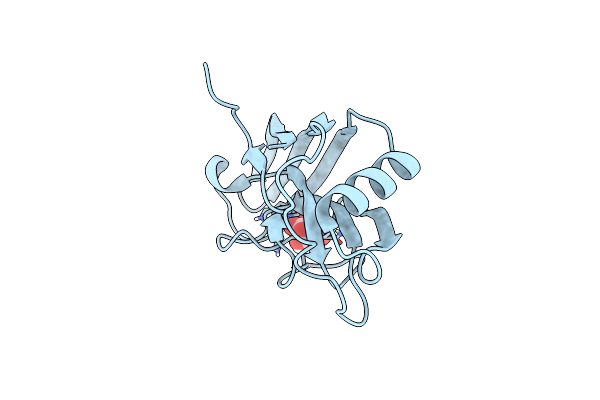 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, TLA |
 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, GLU |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-08-08 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Acetyltransferase Eis From Mycobacterium Tuberculosis In Complex With A Lys-Containing Peptide
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2017-10-25 Classification: SIGNALING PROTEIN |
 |
A Chimeric Microtubule Disruptor With Efficacy On A Taxane Resistant Cell Line
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2010-03-31 Classification: LYASE Ligands: ZN, MS5, FMT |
 |
Highly Potent First Examples Of Dual Aromatase-Steroid Sulfatase Inhibitors Based On A Biphenyl Template
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-02-23 Classification: LYASE Ligands: MS4, ZN |
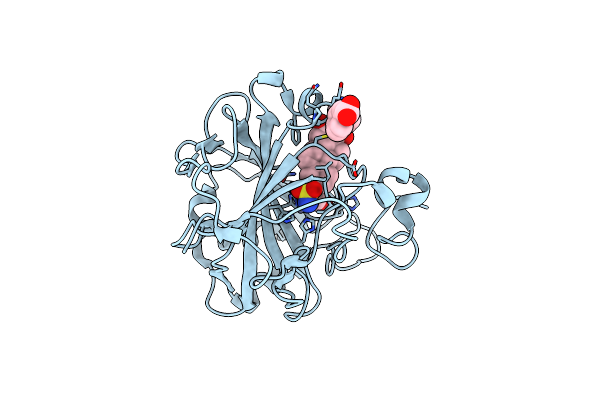 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-01-13 Classification: LYASE Ligands: ZN, CL, POF, MBO, GOL |
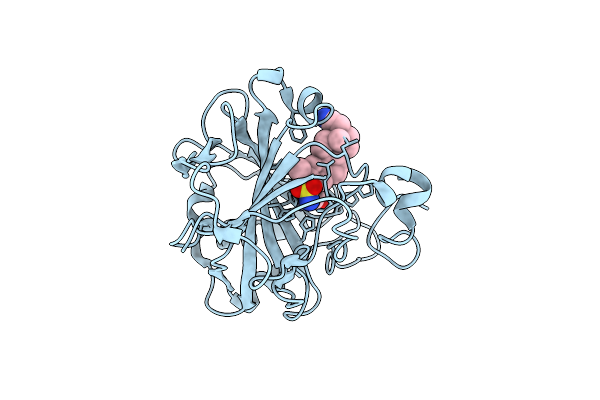 |
Crystal Structure Of The Human Carbonic Anhydrase Ii In Complex With Stx 641 At 1.85 Angstroms Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-10-07 Classification: LYASE Ligands: ZN, CTF, GOL |
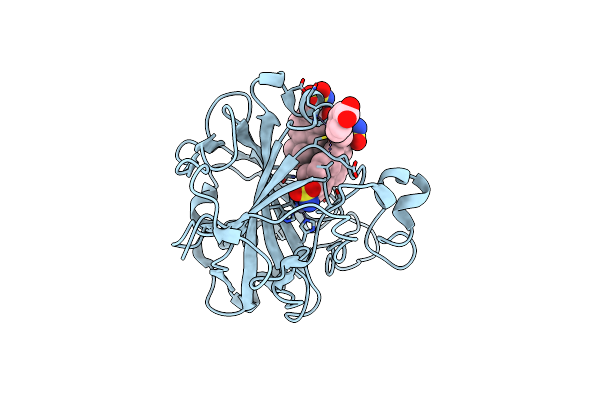 |
Crystal Structure Analysis Of The Human Carbonic Anhydrase Ii In Complex With A 2-Substituted Estradiol Bis-Sulfamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2007-02-06 Classification: LYASE Ligands: ZN, CL, PO1, MBO |
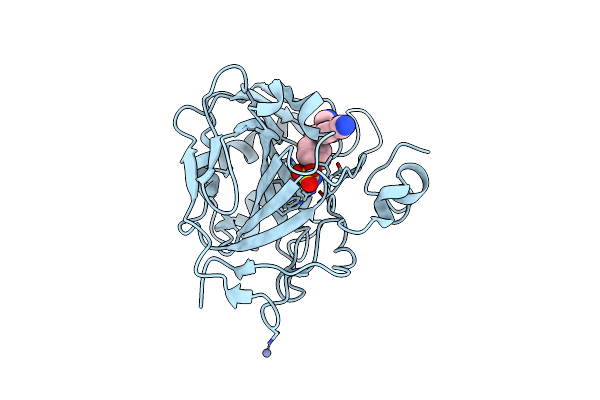 |
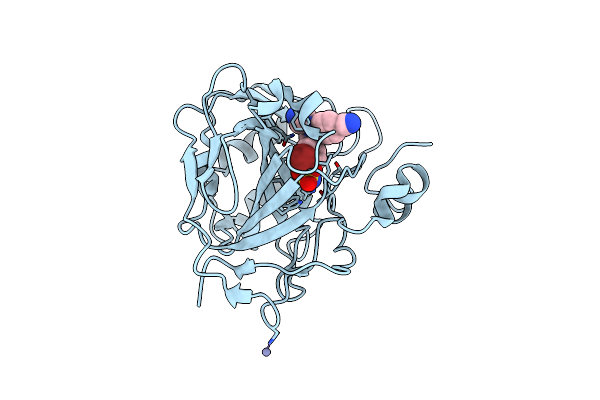
Structure Of Human Carbonic Anhydrase Ii With 4-[4-O-Sulfamoylbenzyl)(4-Cyanophenyl)Amino]-4H-[1,2,4]-Triazole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2005-05-17 Classification: LYASE Ligands: ZN, 4TZ |
 |
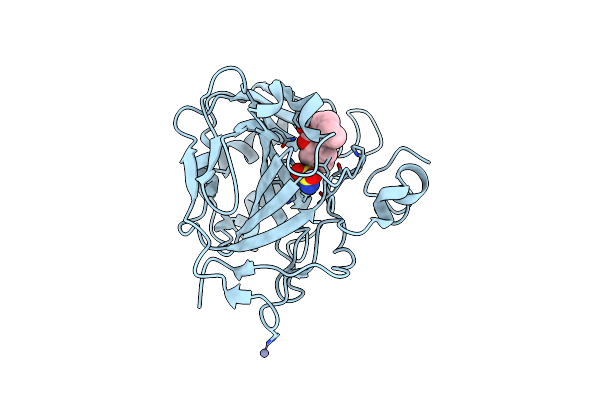
Structure Of Human Carbonic Anhydrase Ii With 4-[(3-Bromo-4-O-Sulfamoylbenzyl)(4-Cyanophenyl)Amino]-4H-[1,2,4]-Triazole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2005-05-17 Classification: LYASE Ligands: ZN, 4TR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-10-05 Classification: LYASE Ligands: ZN, 667 |
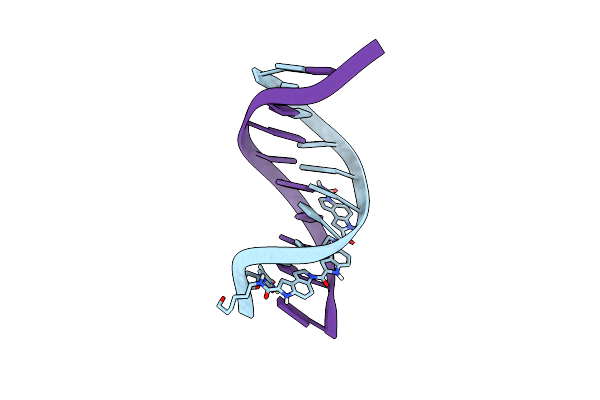 |
Solution Structure Of A Highly Stable Dna Duplex Conjugated To A Minor Groove Binder, Nmr, Minimized Average Structure
|

