Search Count: 56
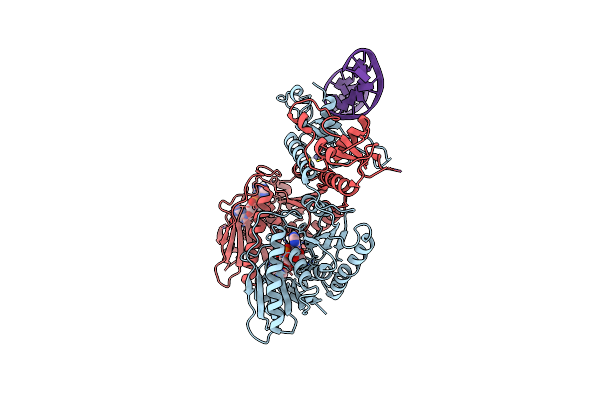 |
Cbass Pseudomonas Syringae Cap5 Tetramer With Dna Duplex And 3'2'-C-Gamp Cyclic Dinucleotide Ligand
Organism: Pseudomonas syringae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: 4UR, ZN |
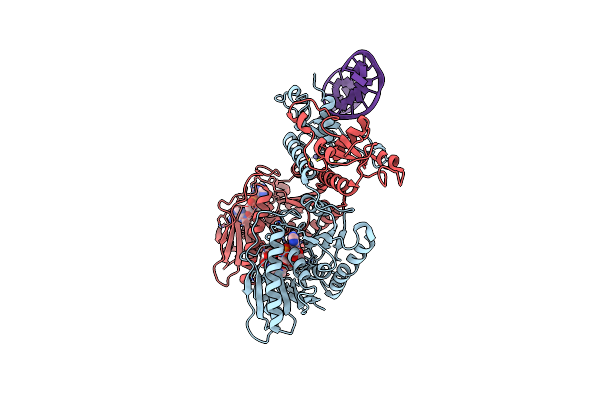 |
Cbass Pseudomonas Syringae Cap5 Tetramer With Dna Duplex And 3'2'-C-Diamp Cyclic Dinucleotide Ligand
Organism: Pseudomonas syringae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: ZN, Y4F, GOL |
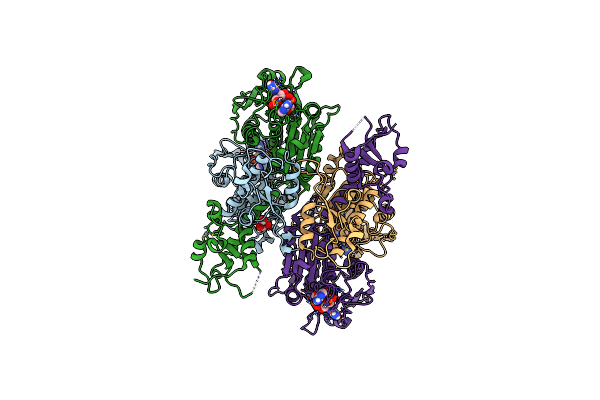 |
Cbass Pseudomonas Syringae Cap5 Tetramer With 3'2'-C-Gamp Cyclic Dinucleotide Ligand (His56Ala Mutant Without Mg2+ Ions)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: IMMUNE SYSTEM Ligands: ZN, 4UR, GOL |
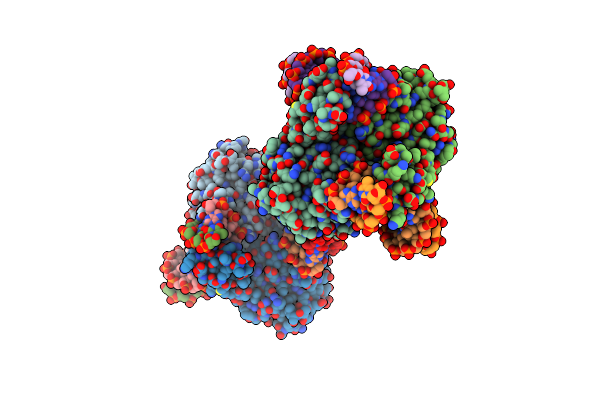 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrates
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: EPE, SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtatac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtaaac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtttac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae In The Absence Of A Ligand (Apo Form Dimer)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae As An Activated Tetramer With The Cyclic Dinucleotide 3'2'-C-Diamp Ligand (1 Tetramer In The Au)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: Y4F, GOL, RWB, ZN, MG, P6G |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae As An Activated Tetramer With The Cyclic Dinucleotide 3'2'-C-Diamp Ligand (3 Tetramers In The Au)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: ZN, MG, Y4F |
 |
Structure Of Cbass Cap5 From Pseudomonas Syringae As An Activated Tetramer With The Cyclic Dinucleotide 3'2'-C-Dgamp Ligand (2 Tetramers In The Au)
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-02-07 Classification: IMMUNE SYSTEM Ligands: 4UR, ZN, MG, PGE, PEG, GOL |
 |
Structure Of Nsp14 N7-Methyltransferase Domain Fused With Telsam Bound To Sgc0946
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-07-26 Classification: VIRAL PROTEIN Ligands: ZN, AW2 |
 |
Structure Of Nsp14 N7-Methyltransferase Domain Fused With Telsam Bound To Sgc8158
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-07-26 Classification: VIRAL PROTEIN Ligands: ZN, EOH, MJ7, K |
 |
Structure Of Nsp14 N7-Methyltransferase Domain Fused With Telsam Bound To Sam
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-09-07 Classification: VIRAL PROTEIN Ligands: ZN, SAM |
 |
Structure Of Nsp14 N7-Methyltransferase Domain Fused With Telsam Bound To Sah
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-07 Classification: VIRAL PROTEIN Ligands: SAH, ZN |
 |
Structure Of Nsp14 N7-Methyltransferase Domain Fused With Telsam Bound To Sinefungin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-09-07 Classification: VIRAL PROTEIN Ligands: SFG, ZN, PEG, EOH, MOH |
 |
Structure Of Human Ck2 Alpha Kinase (Catalytic Subunit) With The Inhibitor 108600.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, GOL, ON6 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2021-06-30 Classification: TRANSFERASE/DNA Ligands: DCP, CA, GOL, PEG |
 |
Human Primpol Misinserting Datp Opposite The 8-Oxoguanine Lesion (3'-End Base Of The Primer Strand Is 2',3'-Dideoxy-Terminated).
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2021-06-30 Classification: TRANSFERASE/DNA Ligands: DTP, CA, GOL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-06-30 Classification: TRANSFERASE/DNA Ligands: DTP, CA, GOL |

