Search Count: 10
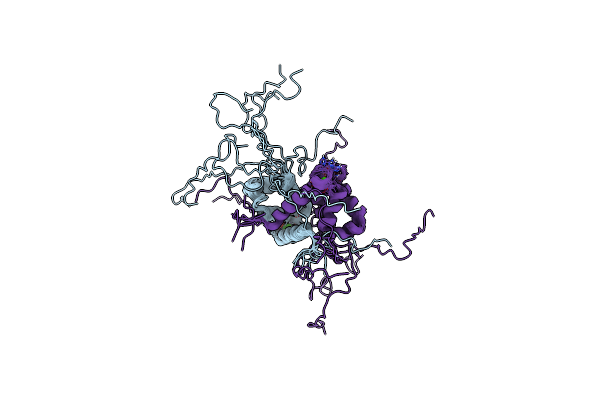 |
Evolution Avoids A Pathological Stabilizing Interaction In The Immune Protein S100A9
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-10-26 Classification: SIGNALING PROTEIN Ligands: CA |
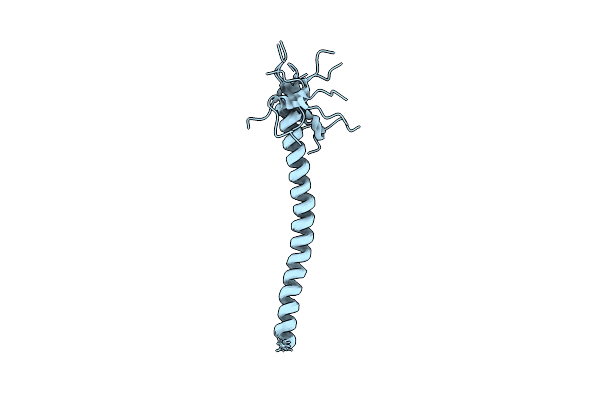 |
Structure Of The Type Iva Major Pilin From The Electrically Conductive Bacterial Nanowires Of Geobacter Sulfurreducens
Organism: Geobacter sulfurreducens
Method: SOLUTION NMR Release Date: 2013-08-28 Classification: Cell Adhesion, Structural Protein, Electron Transport |
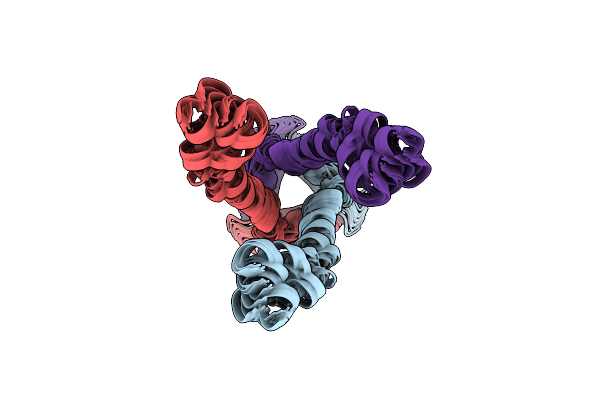 |
Independently Verified Structure Of Gp41-M-Mat, A Membrane Associated Mper Trimer From Hiv-1 Gp41
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2013-05-15 Classification: VIRAL PROTEIN |
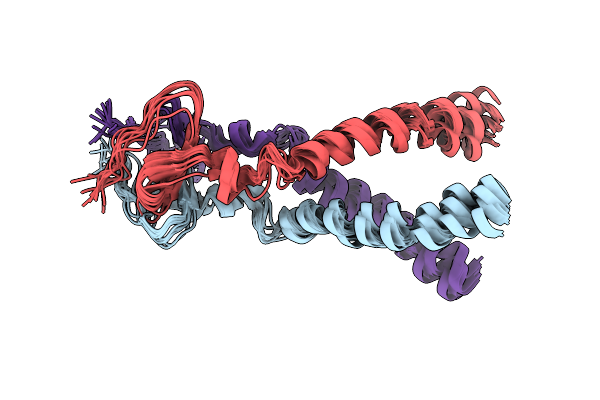 |
Structure Of Gp41-M-Mat, A Membrane Associated Mper Trimer From Hiv-1 Gp41.
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2013-02-06 Classification: VIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-07-29 Classification: CHAPERONE |
 |
Yeast Hsp82 N-Terminal Domain-Geldanamycin Complex: Effects Of Mutants 98-99 Ks-Aa
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-07-29 Classification: CHAPERONE/Antibiotic Ligands: GDM, GOL |
 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-04-03 Classification: CHAPERONE Ligands: RDA, PG4, 1PE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-02-06 Classification: CHAPERONE Ligands: RDA, GOL |
 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2006-10-24 Classification: CHAPERONE Ligands: GDM, PG4, 1PE |
 |
Grp94 N-Terminal Domain Bound To Geldanamycin: Effects Of Mutants 168-169 Ks-Aa
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-10-03 Classification: CHAPERONE Ligands: GDM, PG4, PGE |

