Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
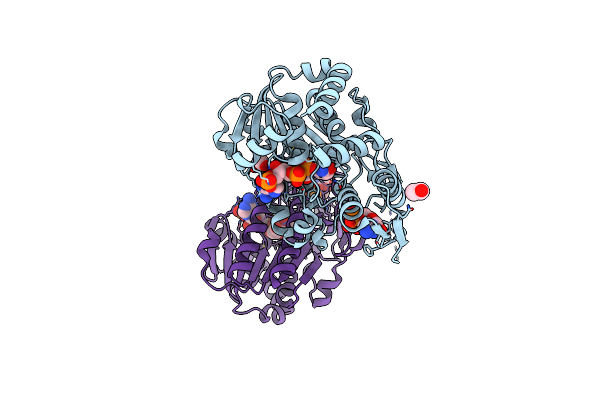 |
X-Ray Structure Of The Nadp-Dependent Reductase From Campylobacter Jejuni Responsible For The Synthesis Of Cdp-Glucitol In The Presence Of Cdp And Nadp
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-20 Classification: OXIDOREDUCTASE Ligands: NAP, CDP, CL, EDO |
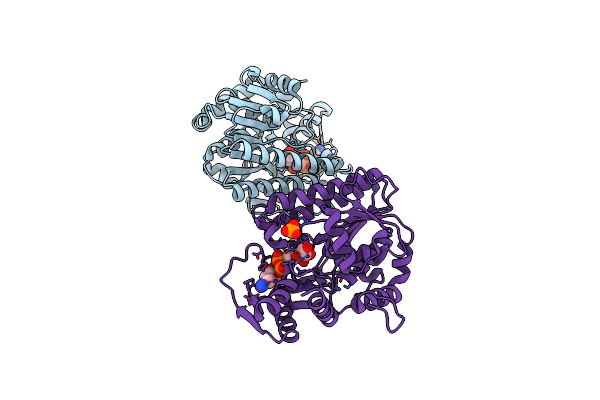 |
X-Ray Structure Of The Nadp-Dependent Reductase From Campylobacter Jejuni Responsible For The Synthesis Of Cdp-Glucitol In The Presence Of Cdp-Glucitol
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-20 Classification: OXIDOREDUCTASE Ligands: YCX, CL, PO4, NA |
 |
X-Ray Structure Of The Gdp-6-Deoxy-4-Keto-D-Lyxo-Heptose-4-Reductase From Campylobacter Jejuni Hs:15
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-11-09 Classification: BIOSYNTHETIC PROTEIN, OXIDOREDUCTASE Ligands: GDP, NDP, EDO |
 |
X-Ray Crystal Structure Of Gdp-D-Glycero-D-Manno-Heptose 4,6-Dehydratase From Campylobacter Jejuni
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-06 Classification: OXIDOREDUCTASE Ligands: NAD, GDP, EDO |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3-Epimerase From Campylobacter Jejuni, Serotype Hs:23/36
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-06-29 Classification: ISOMERASE Ligands: GDP, K, EDO, NA |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3,5-Epimerase From Campylobacter Jejuni, Serotype Hs:42
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-29 Classification: ISOMERASE Ligands: GDP, CL, EDO |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3-Epimerase From Campylobacter Jejuni, Serotype Hs:3
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-06-22 Classification: ISOMERASE Ligands: GDP, CL, TMA |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3,5-Epimerase From Campylobacter Jejuni, Serotype Hs:15
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-22 Classification: ISOMERASE Ligands: GDP, CL |
 |
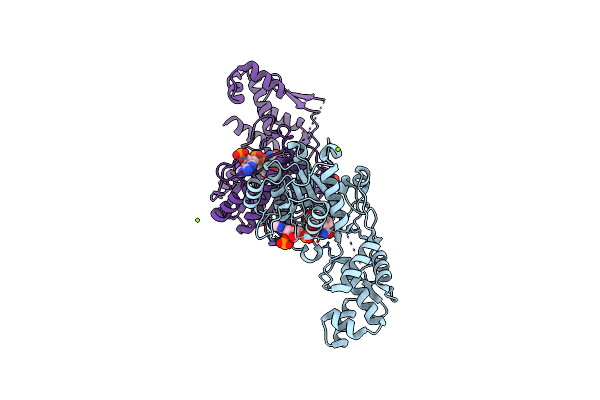
Amycolatopsis Sp. T-1-60 N-Succinylamino Acid Racemase/O-Succinylbenzoate Synthase R266Q Mutant In Complex With N-Succinylphenylglycine
Organism: Amycolatopsis sp.
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-10-06 Classification: ISOMERASE Ligands: 8JI, MG, 1PE |
 |
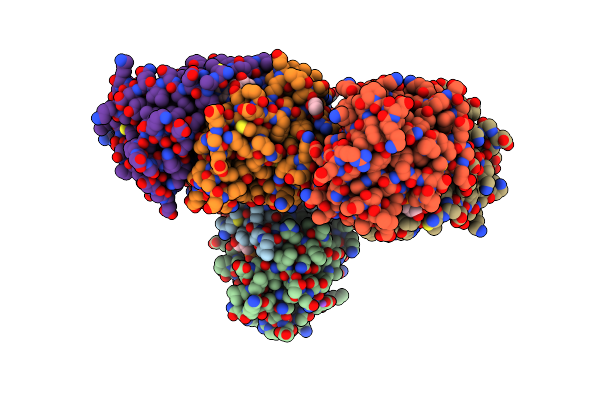
Crystal Structure Of Cj1428, A Gdp-D-Glycero-L-Gluco-Heptose Synthase From Campylobacter Jejuni In The Presence Of Nadph
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-03-24 Classification: OXIDOREDUCTASE Ligands: NAP, MG, EDO |
 |
X-Ray Structure Of Cj1430 In The Presence Of Gdp, A Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3,5-Epimerase From Campylobacter Jejuni
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-24 Classification: ISOMERASE Ligands: GDP, NA, EDO |
 |
Crystal Structure Of Cj1430 In The Presence Of Gdp-D-Glycero-L-Gluco-Heptose, A Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3,5-Epimerase From Campylobacter Jejuni
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-03-24 Classification: ISOMERASE Ligands: YO7, EDO |
 |
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-01-13 Classification: OXIDOREDUCTASE Ligands: UPG, NAD |
 |
Crystal Structure Of 3-Keto-D-Glucoside 4-Epimerase, Ycjr, From E. Coli, Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-06-10 Classification: ISOMERASE Ligands: MN, EDO |
 |
Crystal Structure Of Cj1427, An Essential Nad-Dependent Dehydrogenase From Campylobacter Jejuni, In The Presence Of Nadh And Gdp
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: NAI, GDP, EDO, CL, TMA, NA |
 |
X-Ray Structure Of The Cj1427 In The Presence Of Nadh And Gdp-D-Glycero-D-Mannoheptose, An Essential Nad-Dependent Dehydrogenase From Campylobacter Jejuni
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE Ligands: NAI, GZ0 |
 |
Organism: Novosphingobium sp. (strain ka1)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-07-31 Classification: ISOMERASE Ligands: NQM, CL |
 |
Organism: Novosphingobium sp. (strain ka1)
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-07-31 Classification: ISOMERASE Ligands: CL, CA, NA |
 |
Organism: Novosphingobium sp. (strain ka1)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-07-31 Classification: ISOMERASE Ligands: CL |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-04-24 Classification: LYASE Ligands: ZN, CL |