Search Count: 16
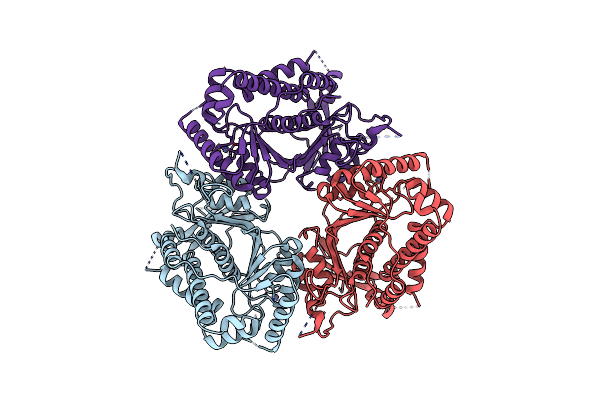 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Trimer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
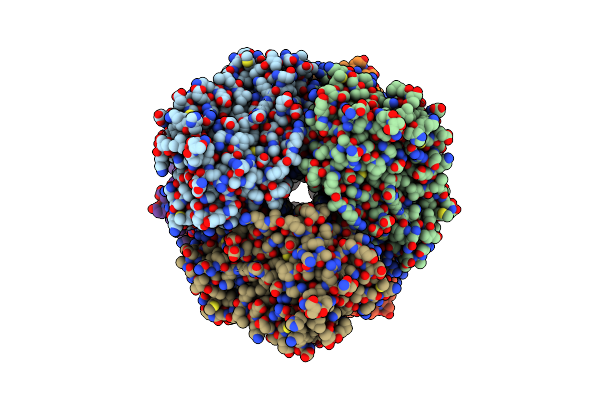 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
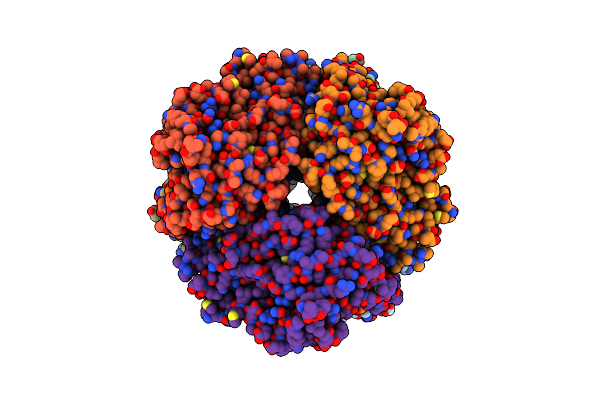 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With Dctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, DCP |
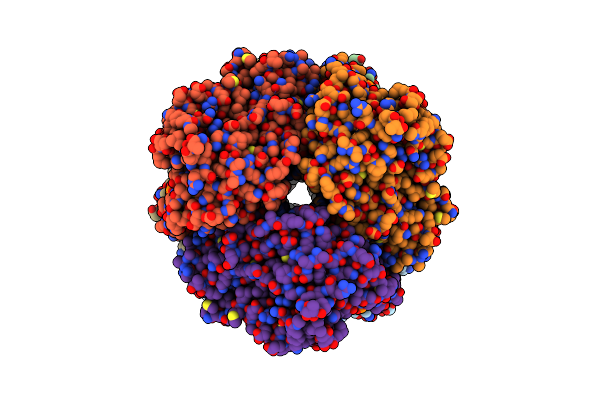 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With 5Mdctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, A1I2I |
 |
Organism: Thermococcus kodakarensis
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: ISOMERASE |
 |
Crystal Structure Of Ritonavir Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-01 Classification: HYDROLASE Ligands: RIT, CPS, EDO |
 |
Crystal Structure Of Lopinavir Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-02-01 Classification: HYDROLASE Ligands: AB1, CPS |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: NAG, EDO |
 |
Crystal Structure Of Plasmodium Falciparum Histo-Aspartic Protease (Hap) Zymogen (Form 1)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-27 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Crystal Structure Of Plasmodium Falciparum Histo-Aspartic Protease (Hap) Zymogen (Form 2)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-05-27 Classification: HYDROLASE Ligands: PEG, GOL |
 |
Crystal Structure Of Plasmodium Falciparum Histo-Aspartic Protease (Hap) Zymogen (Form 3)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-05-27 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Kni-10343 Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-07-11 Classification: HYDROLASE Ligands: 8V9, CPS, GOL, PO4 |
 |
Crystal Structure Of Kni-10743 Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-07-11 Classification: HYDROLASE Ligands: 8VC, GOL, EDO, CPS |
 |
Crystal Structure Of Kni-10333 Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-07-11 Classification: HYDROLASE Ligands: 8VO, CPS, GOL |
 |
Crystal Structure Of Kni-10395 Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-07-11 Classification: HYDROLASE Ligands: K95, CPS, NA |
 |
Crystal Structure Of Kni-10742 Bound Plasmepsin Ii (Pmii) From Plasmodium Falciparum
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-07-11 Classification: HYDROLASE Ligands: CPS, 8VF, NA |

