Search Count: 17
 |
Organism: Lactococcus phage tp901-1
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-02-28 Classification: DNA BINDING PROTEIN |
 |
Organism: Ignisphaera aggregans (strain dsm 17230 / jcm 13409 / aq1.s1)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2021-09-29 Classification: HYDROLASE Ligands: CA, HEZ, CL |
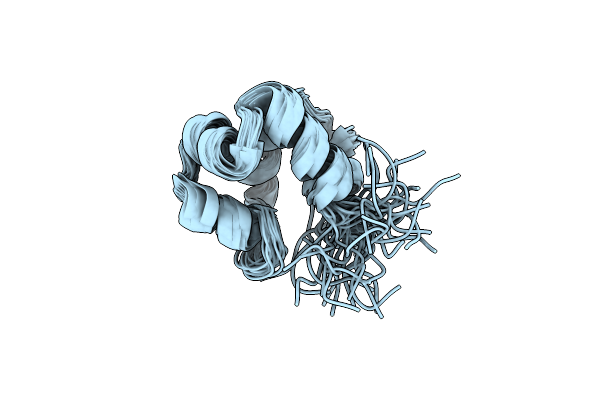 |
Solution Structure Of The Modulator Of Repression (Mor) Of The Temperate Bacteriophage Tp901-1 From Lactococcus Lactis
Organism: Lactococcus phage tp901-1
Method: SOLUTION NMR Release Date: 2020-08-19 Classification: TRANSCRIPTION |
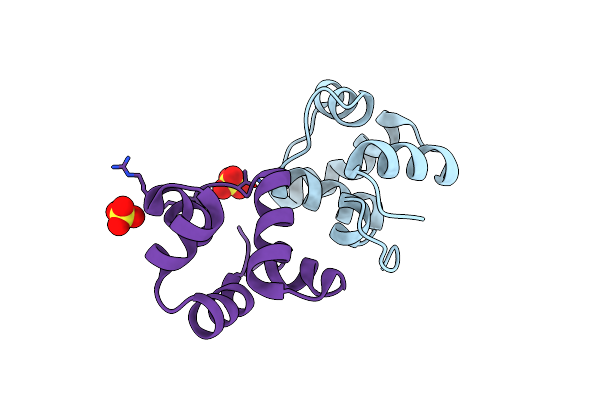 |
Ci-Mor Repressor-Antirepressor Complex Of The Temperate Bacteriophage Tp901-1 From Lactococcus Lactis
Organism: Lactococcus phage tp901-1
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-08-19 Classification: VIRAL PROTEIN Ligands: SO4 |
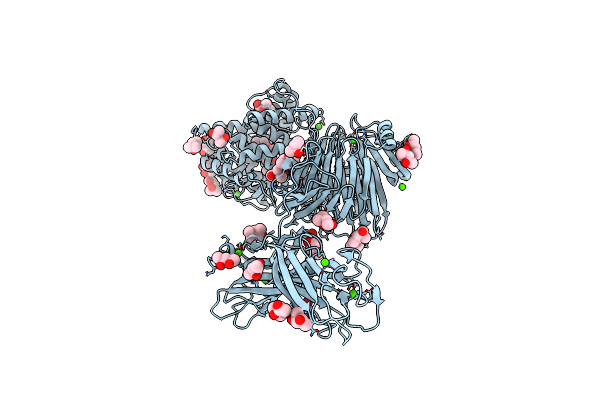 |
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-05 Classification: LYASE Ligands: CA, CL, BTB, MRD, MPD |
 |
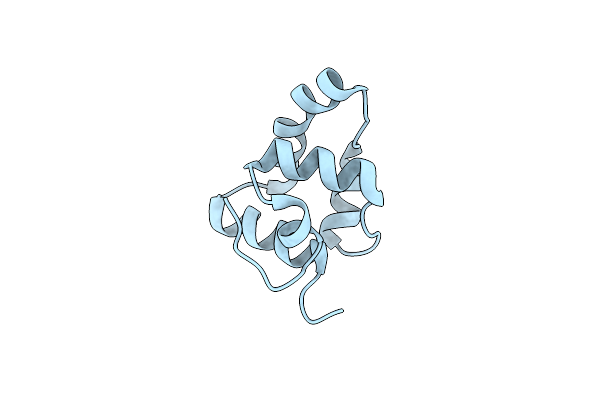
Organism: Lactococcus phage tp901-1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-05-30 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Lactococcus phage
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-07-27 Classification: TRANSCRIPTION |
 |
Organism: Lactococcus phage tp901-1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-12-25 Classification: TRANSCRIPTION |
 |
N-Terminal Domain Of The Ci Repressor From Bacteriophage Tp901-1 In Complex With The Ol2 Operator Half-Site
Organism: Lactococcus phage tp901-1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-12-25 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2011-02-23 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2011-02-23 Classification: CELL ADHESION Ligands: NAG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2011-02-23 Classification: CELL ADHESION Ligands: NAG, EPE, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-08-25 Classification: CELL ADHESION Ligands: NAG, CA |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-11-17 Classification: CELL ADHESION Ligands: NAG, GOL, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-08-26 Classification: CELL ADHESION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-07-29 Classification: CELL ADHESION Ligands: SO4, NAG, GOL |

