Search Count: 21
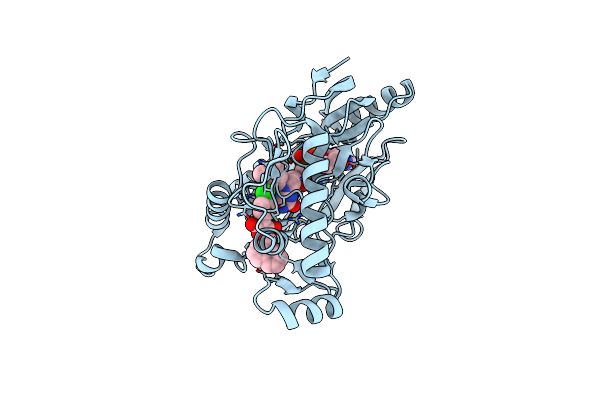 |
Crystal Structure Of E. Coli Mur B Bound To A Naphthyl Tetronic Acid Inihibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2007-06-26 Classification: OXIDOREDUCTASE Ligands: 973, FAD |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 1999-06-08 Classification: HYDROLASE Ligands: ZN, NA |
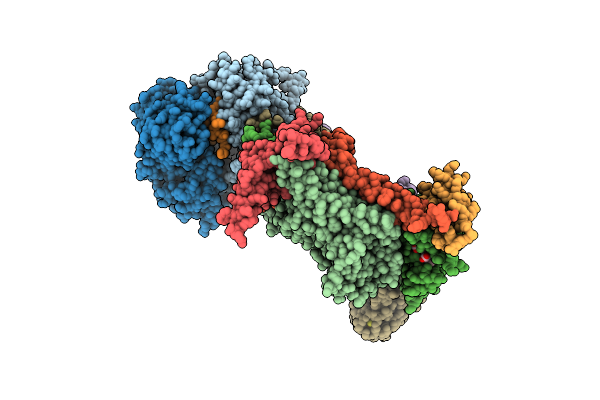 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1999-01-13 Classification: ELECTRON TRANSPORT Ligands: HEM, HEC, FES |
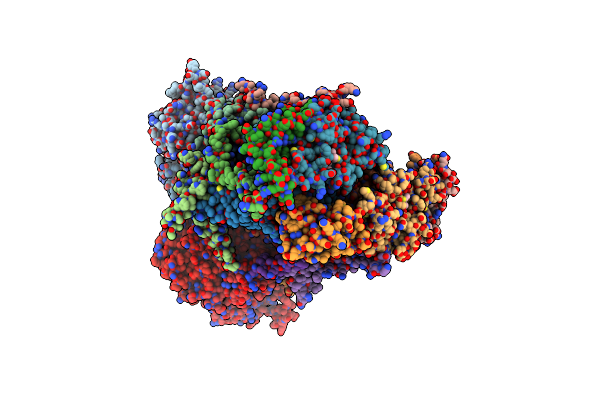 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1999-01-06 Classification: ELECTRON TRANSPORT Ligands: HEM, HEC, FES |
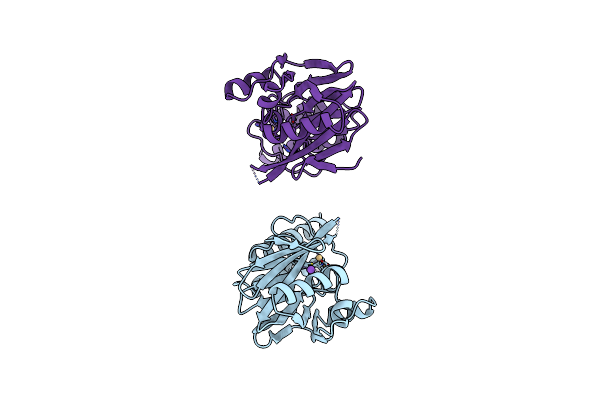 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 1998-01-28 Classification: HYDROLASE Ligands: CD, NA |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 1998-01-28 Classification: HYDROLASE Ligands: ZN, HG, NA |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 1997-08-20 Classification: COMPLEX (TRNA SYNTHETASE/PEPTIDE) Ligands: SO4, HIS |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1997-08-20 Classification: TRNA SYNTHETASE Ligands: SO4, HAM |
 |
The 1.8 Angstrom Crystal Structure Of The Dimeric Peroxisomal Thiolase Of Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-06-16 Classification: THIOLASE Ligands: MRD |
 |
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-01-27 Classification: ISOMERASE Ligands: B12, DCA, GOL |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1997-01-11 Classification: HYDROLASE (BETA-LACTAMASE) Ligands: ZN, NA |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-08-17 Classification: BINDING PROTEIN |
 |
Nature Of The Inactivation Of Elastase By N-Peptidyl-O-Aroyl Hydroxylamine As A Function Of Ph
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-07-10 Classification: COMPLEX (HYDROLASE/INHIBITOR) Ligands: CA, SO4, BAF |
 |
Nature Of The Inactivation Of Elastase By N-Peptidyl-O-Aroyl Hydroxylamine As A Function Of Ph
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1995-07-10 Classification: COMPLEX (HYDROLASE/INHIBITOR) Ligands: CA, BAA |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1994-06-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0Z4, CA, SO4 |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1994-04-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0Z1, CA, SO4, ACY |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 1994-04-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0Z3, CA |
 |
Direct Structure Observation Of An Acyl-Enzyme Intermediate In The Hydrolysis Of An Ester Substrate By Elastase
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1994-04-30 Classification: HYDROLASE(SERINE PROTEINASE) Ligands: CA, SO4 |
 |
Direct Structure Observation Of An Acyl-Enzyme Intermediate In The Hydrolysis Of An Ester Substrate By Elastase
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1994-04-30 Classification: HYDROLASE Ligands: BBL, CA, SO4 |
 |
Structure Of The Mg2+-Bound Form Of Chey And Mechanism Of Phosphoryl Transfer In Bacterial Chemotaxis
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-04-30 Classification: SIGNAL TRANSDUCTION PROTEIN Ligands: MG |

