Search Count: 32
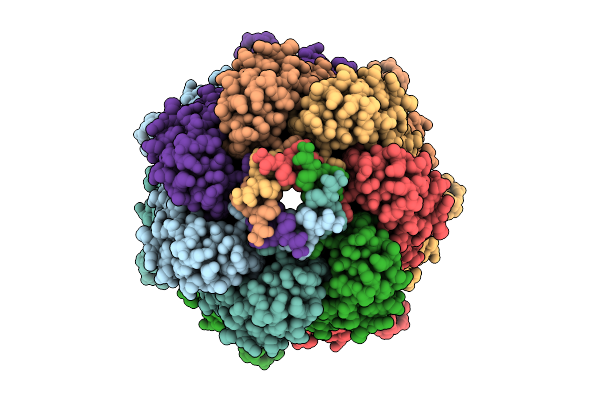 |
Ynai In Its Open Conformation Purified In Ddm Showing Ligand-Filled Pockets
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: D12 |
 |
A Ynai-Mscs Chimera In A Closed Conformation Purified In Ddm With Additional Lipids Showing Ligand-Filled Pore And Pockets
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: D12 |
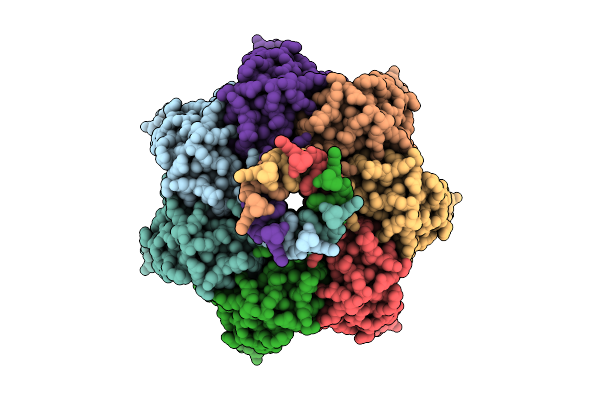 |
A Ynai-Mscs Chimera In An Open Conformation Purified In Ddm Showing Ligand-Filled Pockets
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: D12 |
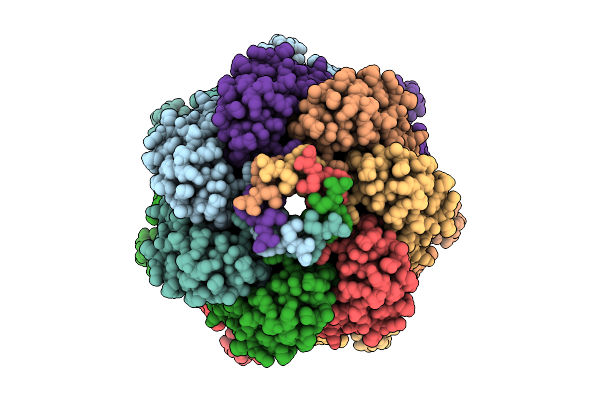 |
Ynai In Closed Conformation Purified In Ddm With Additional Lipids Showing Ligand-Filled Pockets
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: D12 |
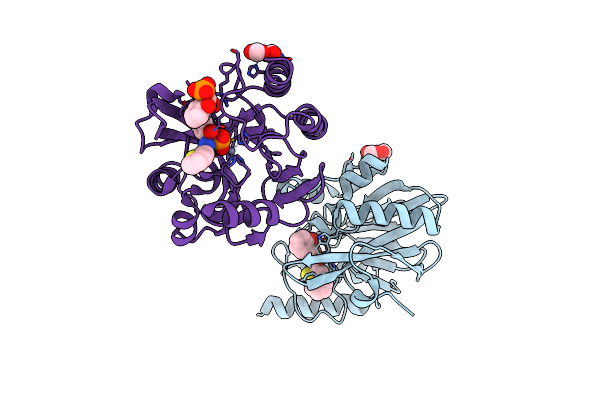 |
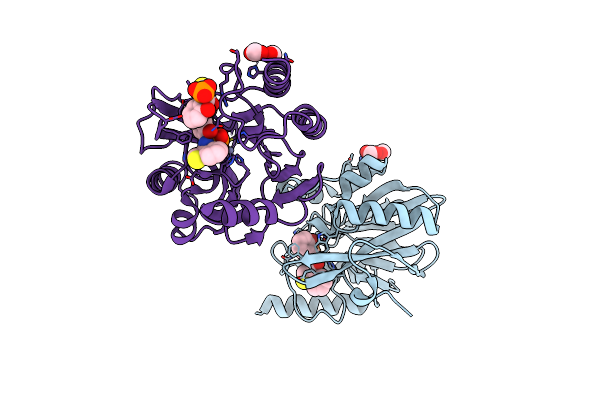
Vim-2 In Complex With Gkv61 (5C) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1IKA, ZN, A1H8T |
 |
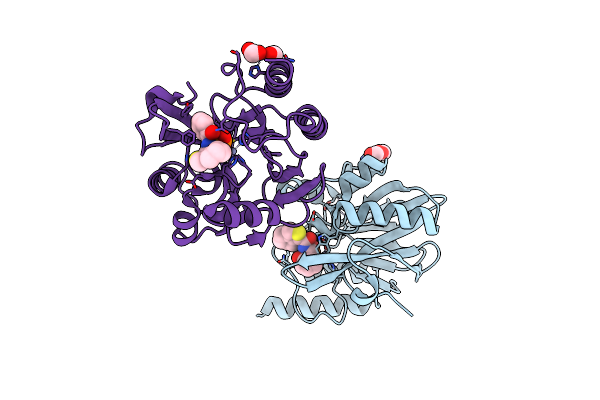
Vim-2 In Complex With Gkv53 (5D) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1IJ9, ZN, A1H8W |
 |
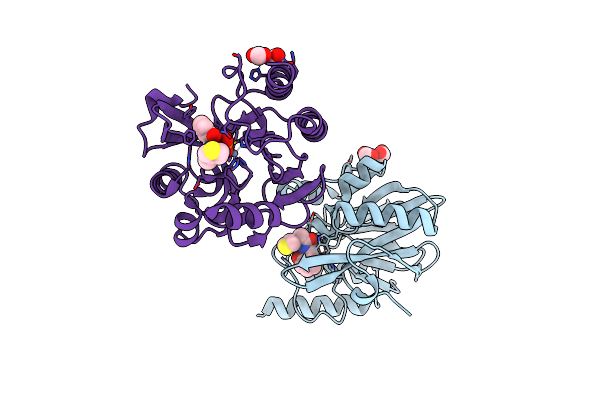
Vim-2 In Complex With Gkv65 (5G) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1H8V, ZN, MG |
 |
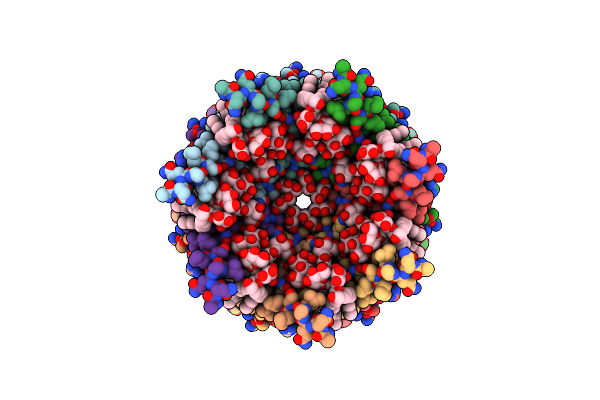
Vim-2 In Complex With Gkv63 (5J) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1H8U, ZN |
 |
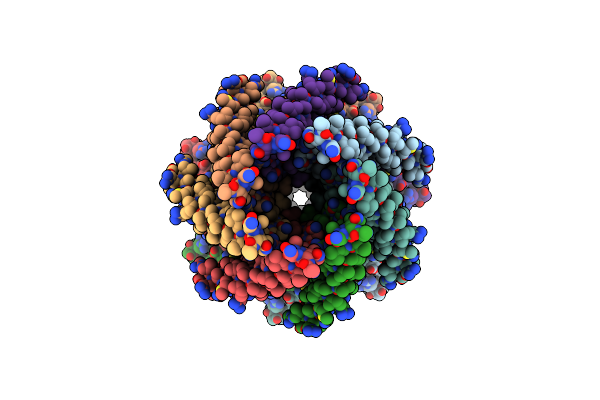
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN Ligands: PEE, LMT, AV0 |
 |
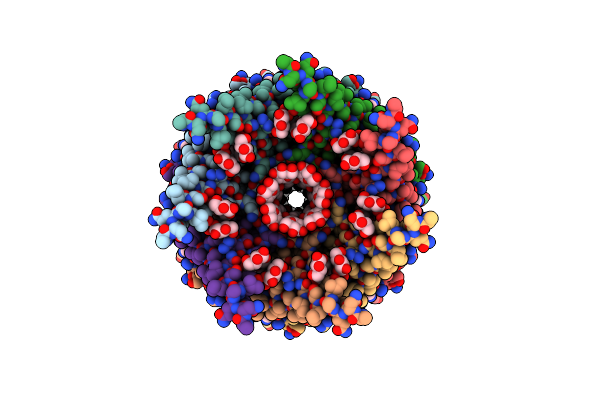
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN |
 |
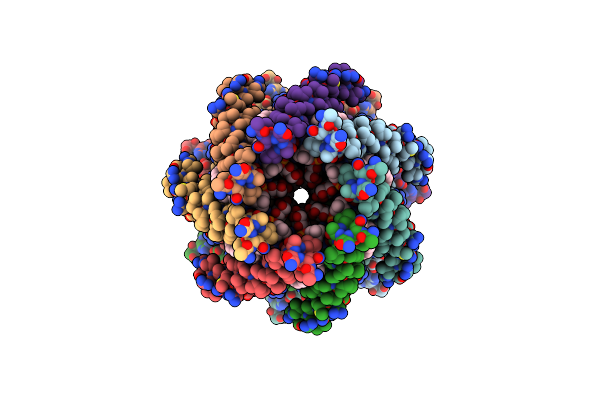
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN Ligands: PEE, LMT |
 |
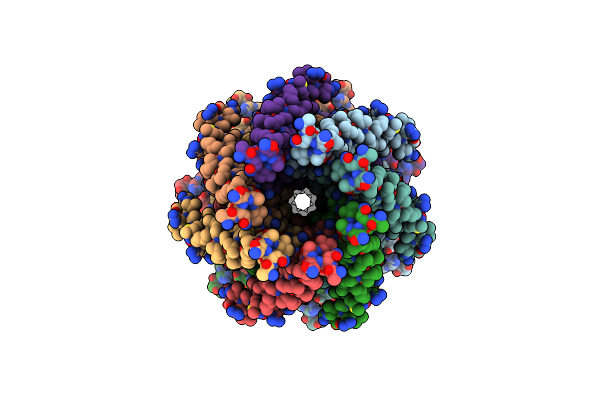
Mechanosensitive Channel Mscs Solubilized With Ddm In Closed Conformation With Added Lipid
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN Ligands: PCW, LMT |
 |
Mechanosensitive Channel Mscs Solubilized With Lmng In Closed Conformation With Added Lipid
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN |
 |
Mechanosensitive Channel Mscs Solubilized With Lmng In Open Conformation With Added Lipid
Organism: escherichia coli se11
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN Ligands: PEE, AV0, LMT |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: MEMBRANE PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN Ligands: ZN, GOL, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-06-24 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Structure Of The Mechanosensitive Channel Mscs Embedded In The Membrane Bilayer
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2019-07-24 Classification: MEMBRANE PROTEIN Ligands: PCW |

