Search Count: 125
 |
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Synthetic construct, Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
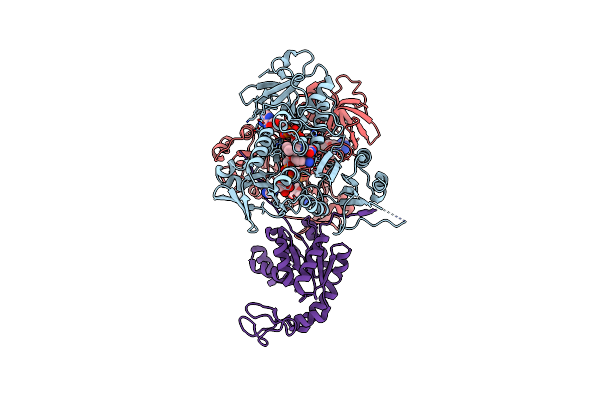
Chromosomal Passenger Complex In Complex With H3T3Ph Nucleosome (Double Occupancy)
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
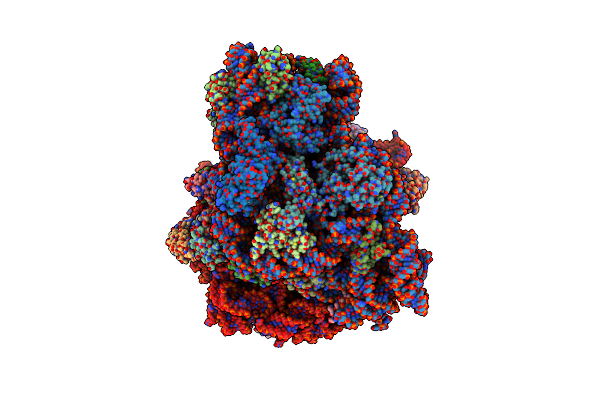
Cryo-Em Structure Of Human Ak2 Bound To Reduced Human Aifm1 (Residues 102-613), Class 3
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: NAD, FAD |
 |
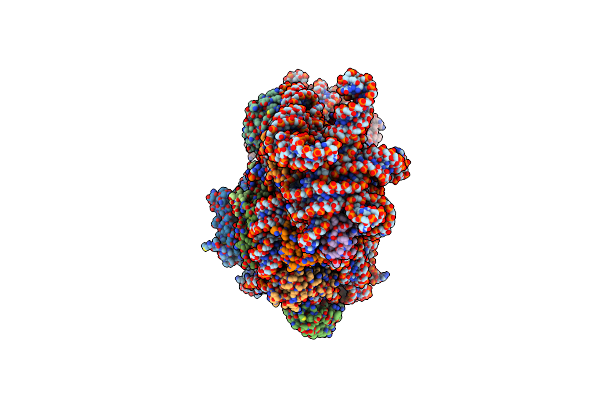
Organism: Escherichia coli, Synthetic construct, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: GENE REGULATION Ligands: ZN, K, MG, PHE |
 |
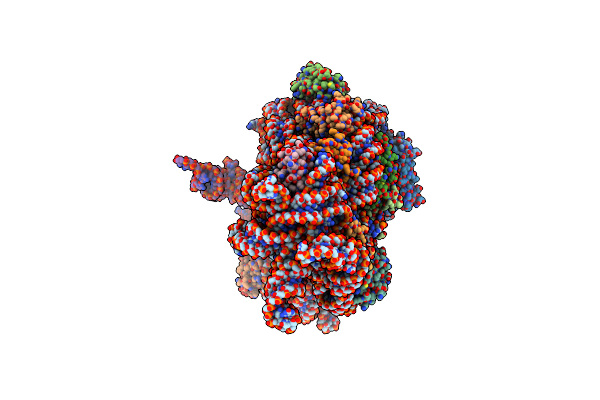
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ZN |
 |
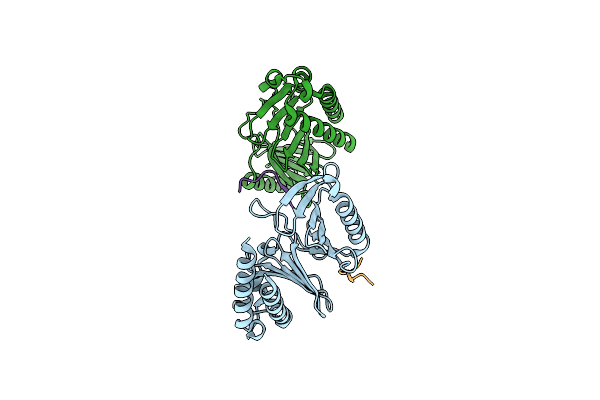
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
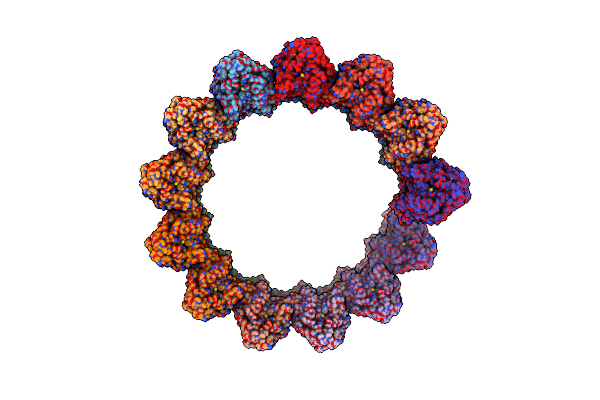 |
Structure Of The Native Microtubule Lattice Nucleated From The Yeast Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE |
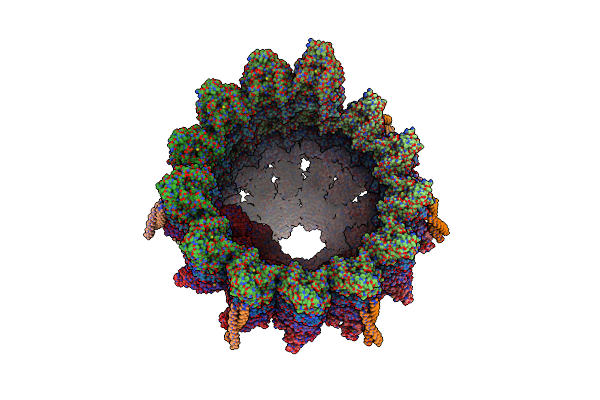 |
Structure Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP, GDP |
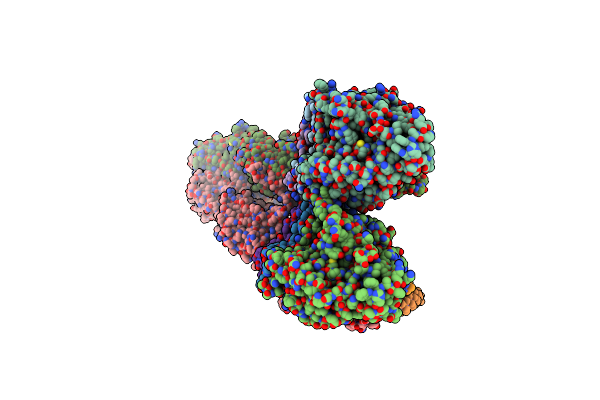 |
Structure Of The Y-Tubulin Small Complex (Ytusc) As Part Of The Native Y-Tubulin Ring Complex (Yturc) Capping Microtubule Minus Ends At The Spindle Pole Body
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: CELL CYCLE Ligands: GTP |

