Search Count: 92
 |
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: BIOSYNTHETIC PROTEIN Ligands: A1EAB, PEG, GOL |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, PEG |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL, A1D9Q |
 |
Organism: Cercospora sp. jnu001
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, GOL |
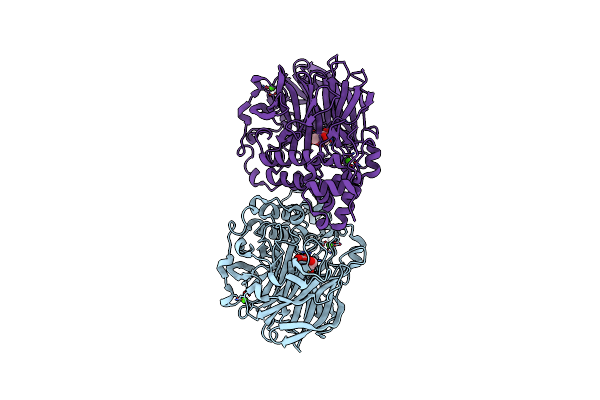 |
Crystal Structure Of Inulosucrase From Lactobacillus Reuteri 121 In Complex With Fructose
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: FRU, CA |
 |
Crystal Structure Of Inulosucrase From Lactobacillus Reuteri 121 Mutant R544W
Organism: Limosilactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, EDO, GOL, 1PE, PEG |
 |
Organism: Aggregatibacter actinomycetemcomitans num4039
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-12-11 Classification: TRANSFERASE |
 |
Organism: Aggregatibacter actinomycetemcomitans num4039
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2024-12-11 Classification: TRANSFERASE |
 |
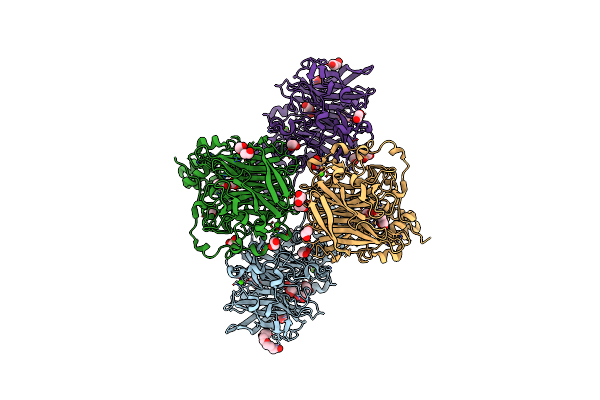
Organism: Aggregatibacter actinomycetemcomitans num4039
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-12-11 Classification: TRANSFERASE Ligands: UDP, PEG, PGE |
 |
Organism: Aggregatibacter actinomycetemcomitans num4039
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2024-12-11 Classification: TRANSFERASE Ligands: UDP, MG |
 |
Organism: Aggregatibacter actinomycetemcomitans num4039
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-12-11 Classification: TRANSFERASE Ligands: GDU, MG |
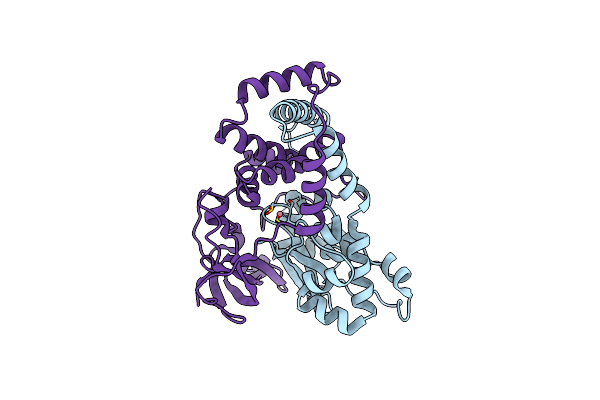 |
Crystal Structure Of Co-Type Nitrile Hydratase Mutant From Pseudomonas Thermophila - L6T
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-14 Classification: LYASE Ligands: CO |
 |
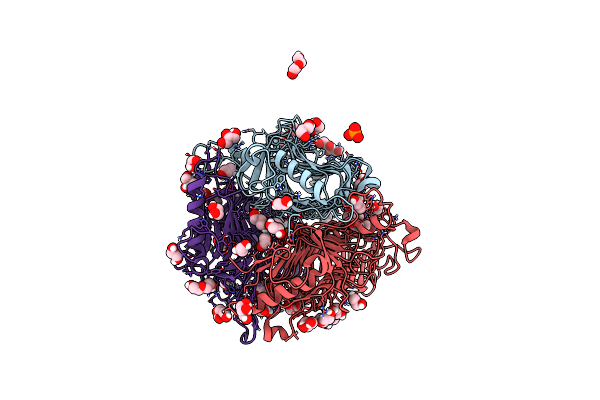 |
Crystal Structure Of Dfa I-Forming Inulin Lyase From Streptomyces Peucetius Subsp. Caesius Atcc 27952
Organism: Streptomyces peucetius subsp. caesius atcc 27952
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2023-12-27 Classification: LYASE Ligands: GOL, EDO, 1PE, PEG, 2PO |
 |
Crystal Structure Of Dfa I-Forming Inulin Lyase From Streptomyces Peucetius Subsp. Caesius Atcc 27952 In Complex With Gf4, Dfa I, And Fructose
Organism: Streptomyces peucetius subsp. caesius atcc 27952
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2023-12-27 Classification: LYASE Ligands: FRU, EDO, MES, PEG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: UDP, GOL |

