Search Count: 15
 |
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FMN, GOL, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-06-15 Classification: MEMBRANE PROTEIN Ligands: V0S, CLR, NAG, NA, MPG |
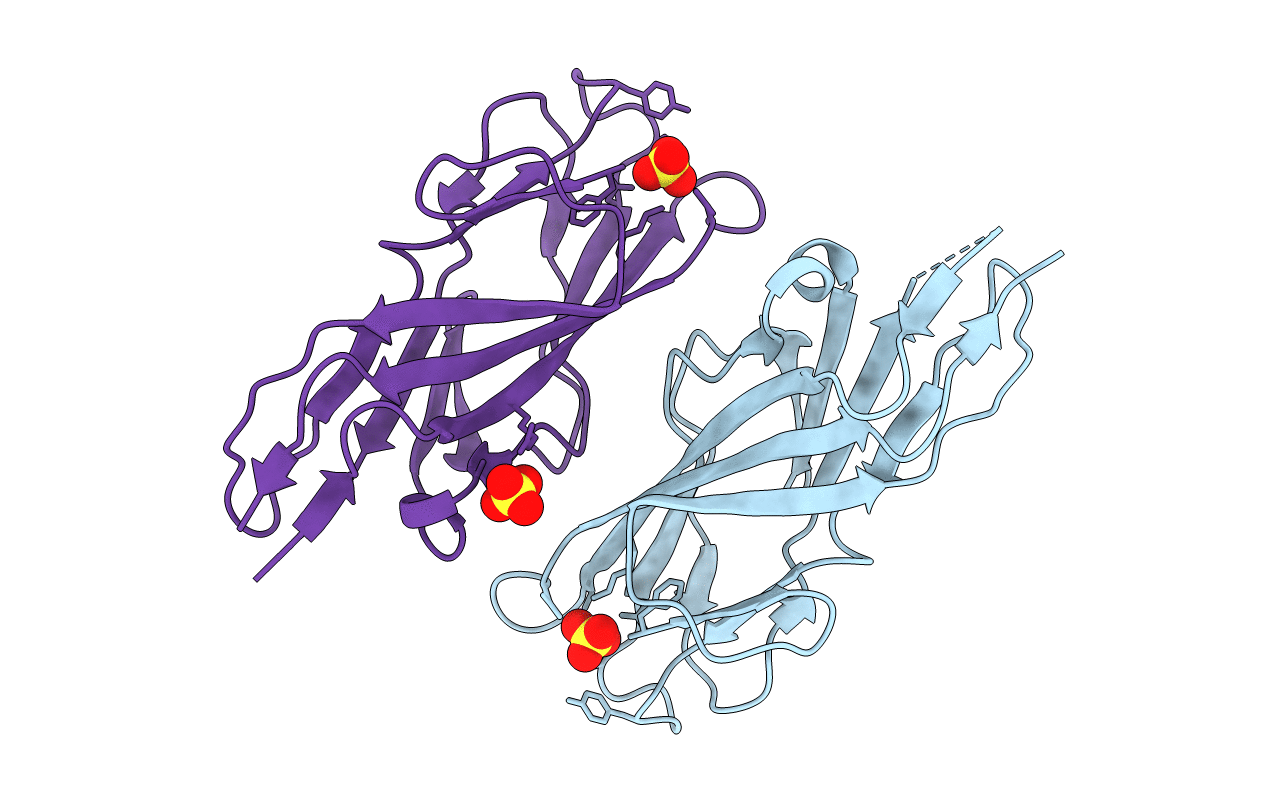 |
Crystal Structure Of Lectin Domain Of F9 Pilus Adhesin Fmlh From E. Coli Uti89
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
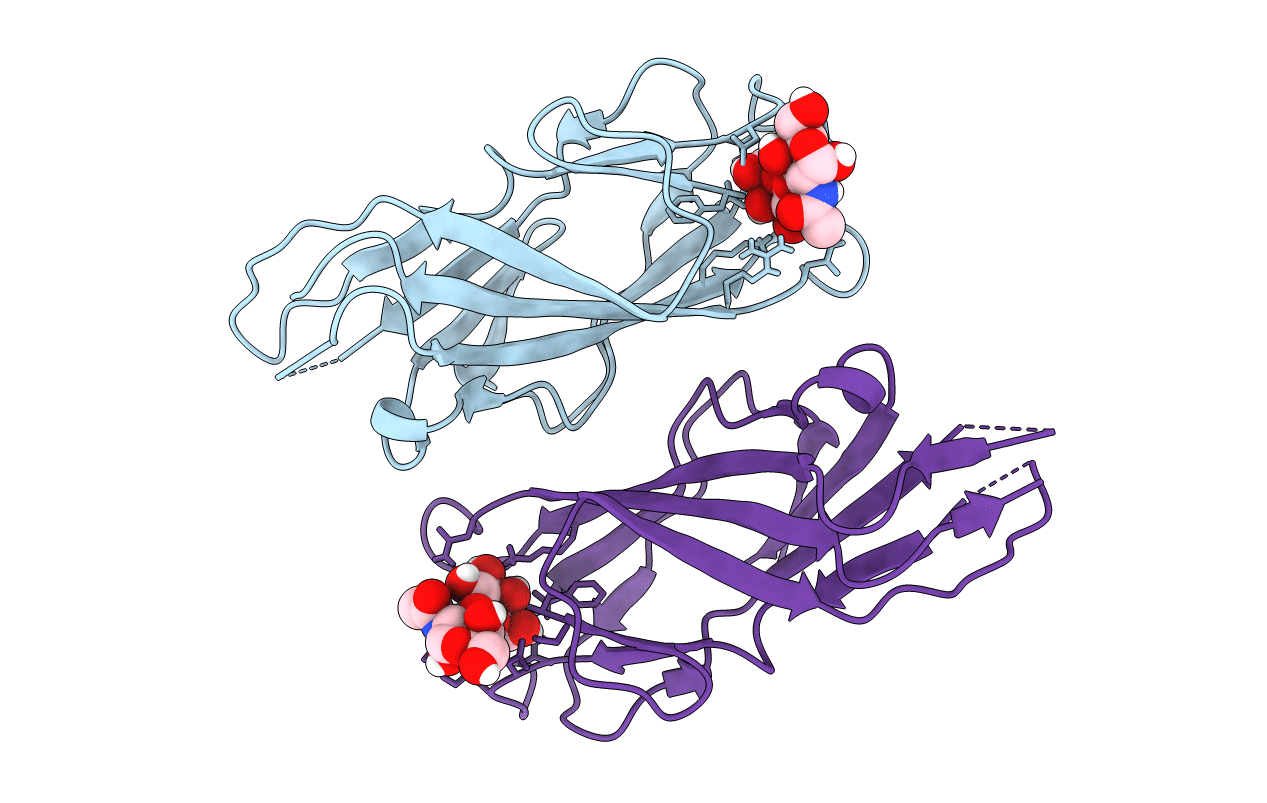 |
F9 Pilus Adhesin Fmlh Lectin Domain From E. Coli Uti89 Co-Crystallized With Tf Antigen
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN |
 |
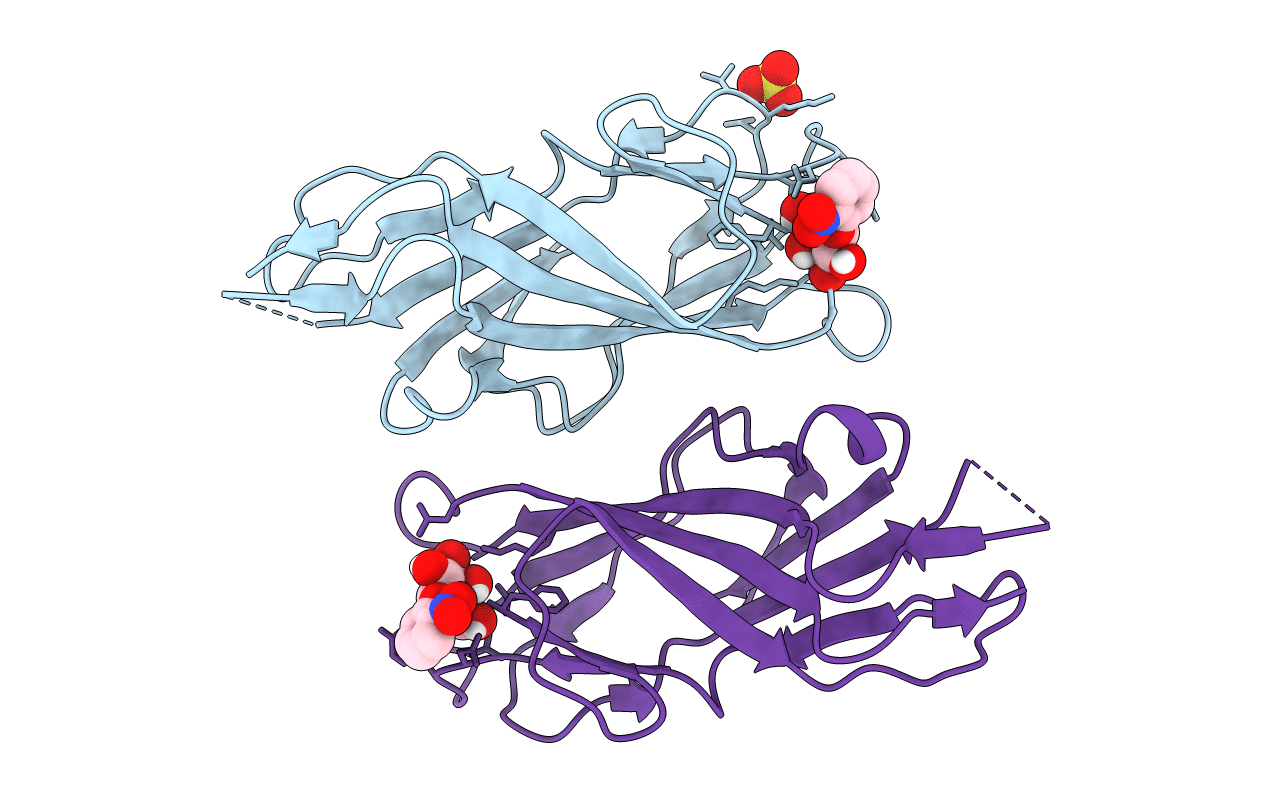
F9 Pilus Adhesin Fmlh Lectin Domain From E. Coli Uti89 Co-Crystallized With O-Nitrophenyl Beta-Galactoside (Onpg)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 145 |
 |
F9 Pilus Adhesin Fmlh Lectin Domain From E. Coli Uti89 Co-Crystallized With O-Nitrophenyl Beta-Galactoside (Onpg)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN Ligands: 145, SO4 |
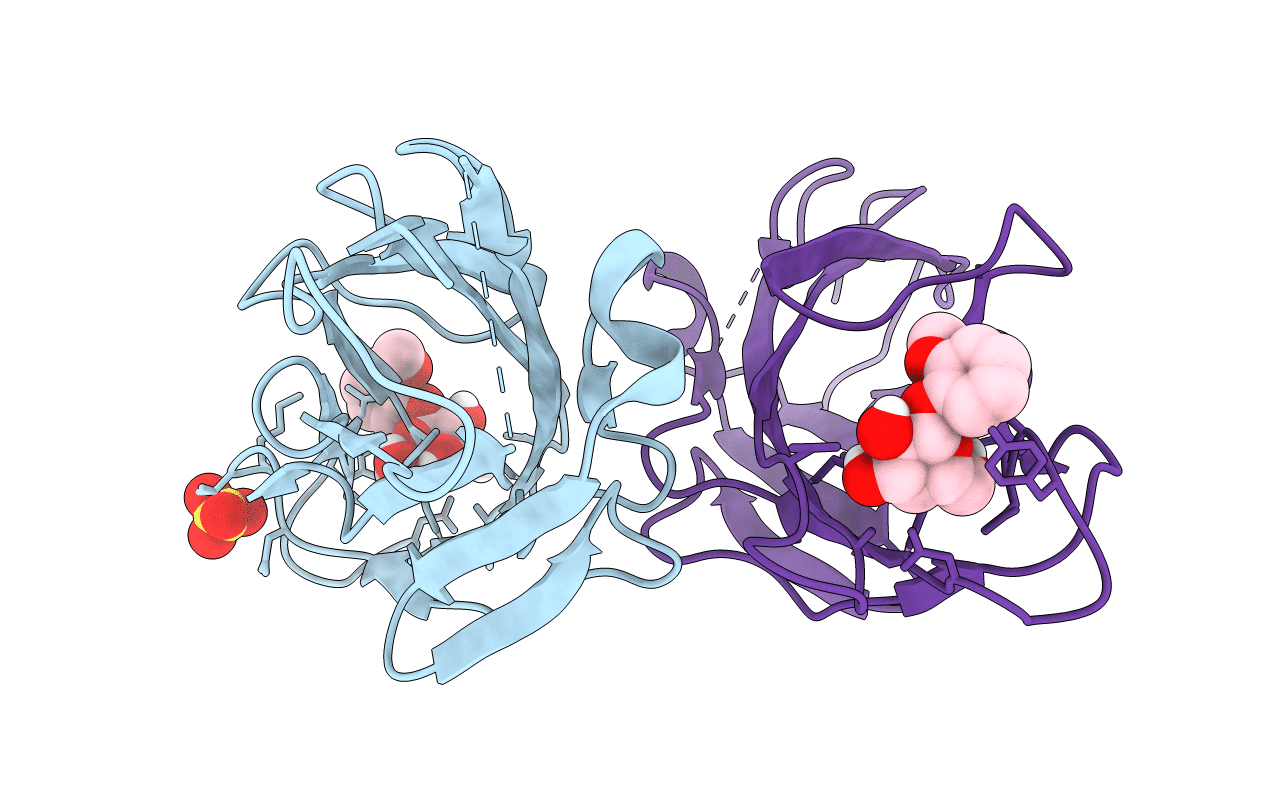 |
F9 Pilus Adhesin Fmlh Lectin Domain From E. Coli Uti89 Co-Crystallized With O-Methoxyphenyl Beta-Galactoside (Ompg)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN Ligands: BQY, SO4 |
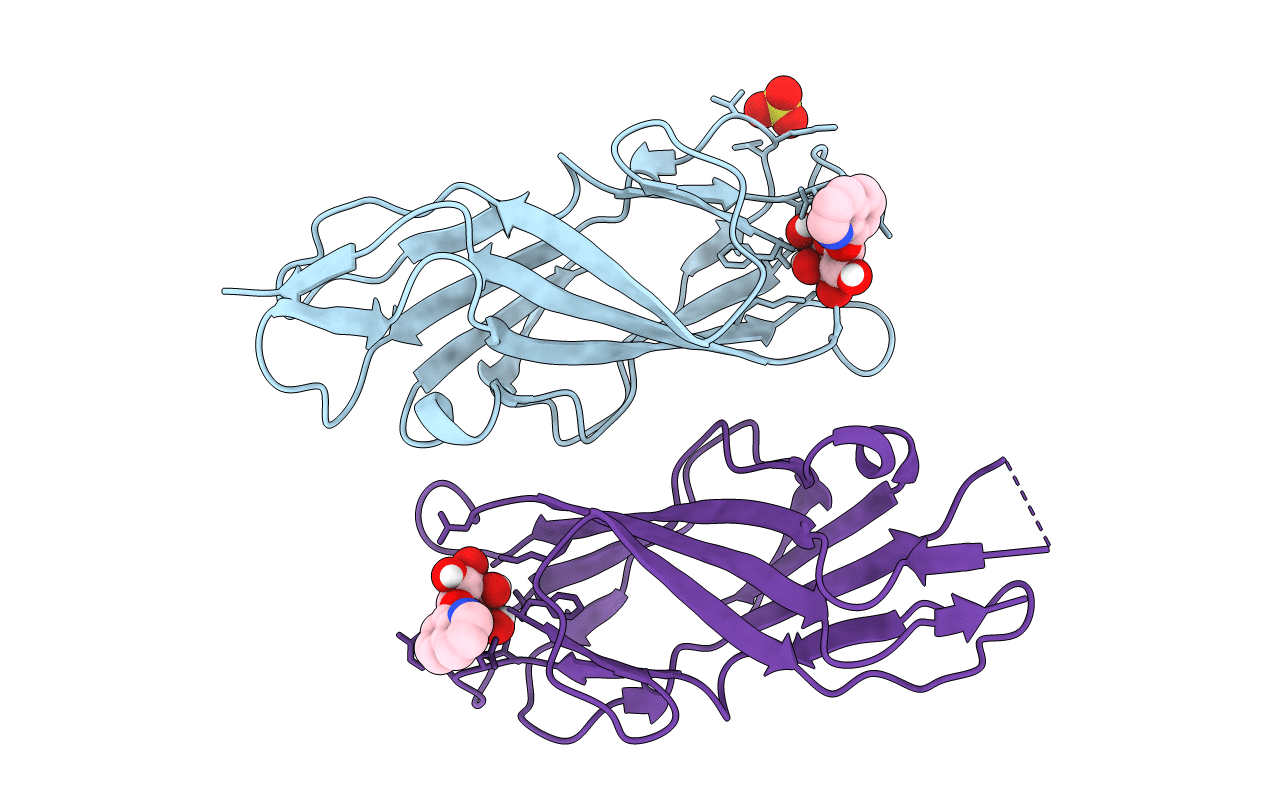 |
F9 Pilus Adhesin Fmlh Lectin Domain From E. Coli Uti89 Co-Crystallized With 8-Hydroxyquinoline Beta-Galactoside
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN Ligands: BS7, SO4 |
 |
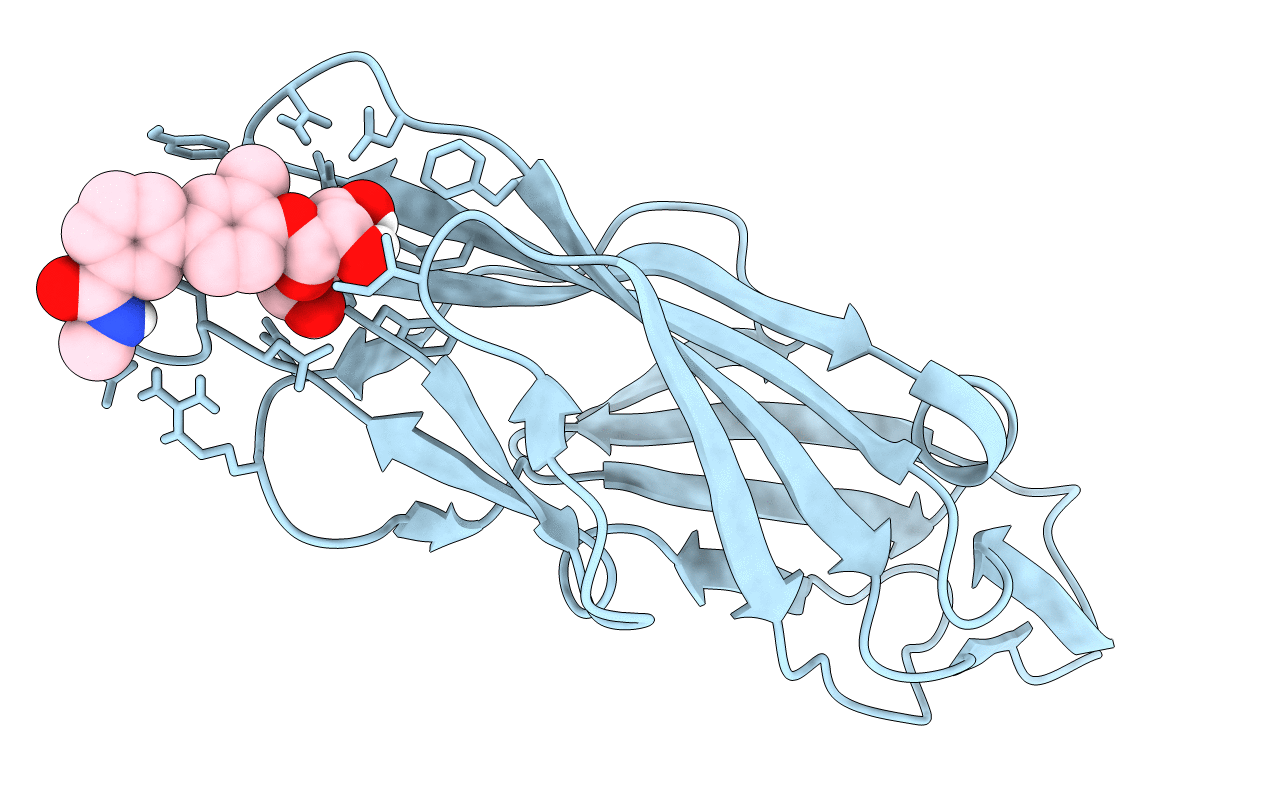
F9 Pilus Adhesin Fmlh Lectin Domain From E. Coli Uti89 Co-Crystallized With Ortho-Biphenyl-2'-Carboxyl N-Acetyl-Beta-Galactosaminoside
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-28 Classification: SUGAR BINDING PROTEIN Ligands: BTG |
 |
Crystal Structure Of Para-Biphenyl-2-Methyl-3'-Methyl Amide Mannoside Bound To Fimh Lectin Domain
Organism: Escherichia coli j96
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-12-07 Classification: SUGAR BINDING PROTEIN Ligands: 5US |
 |
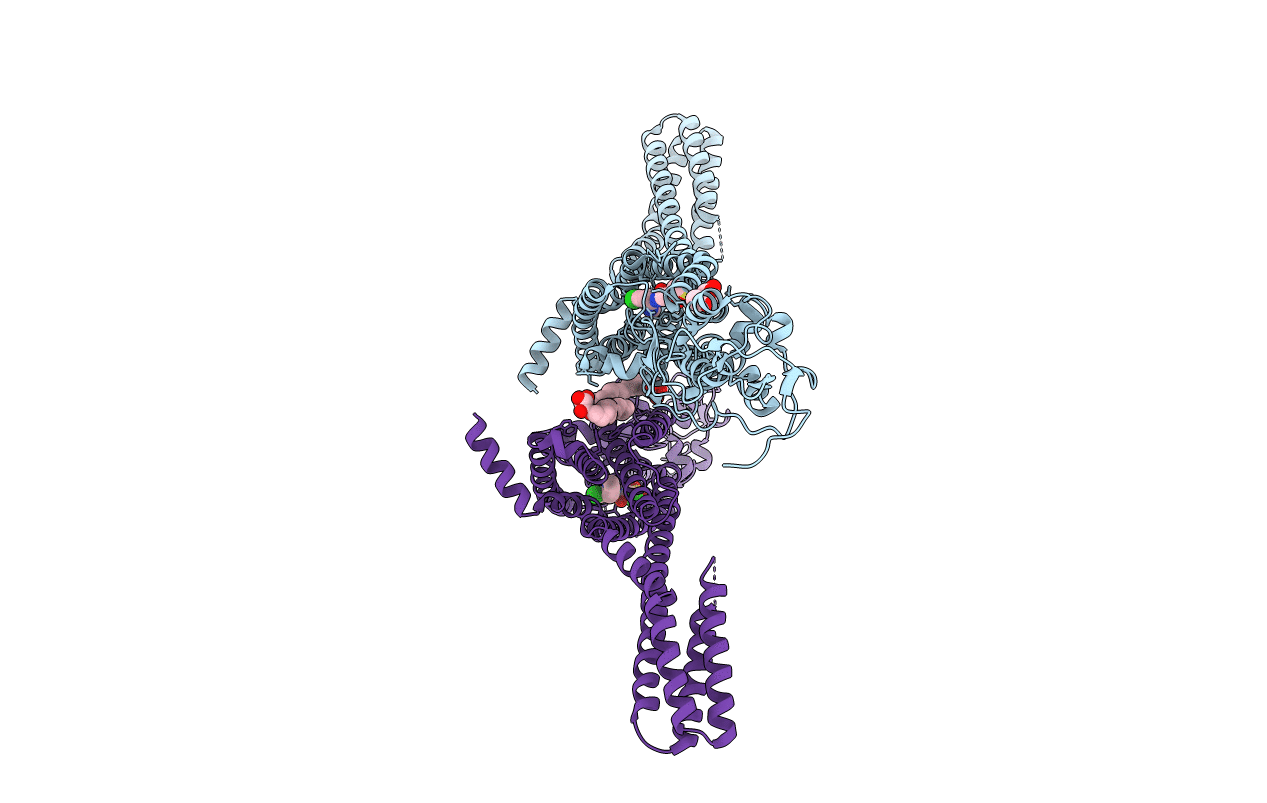
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-07-20 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, NA |
 |
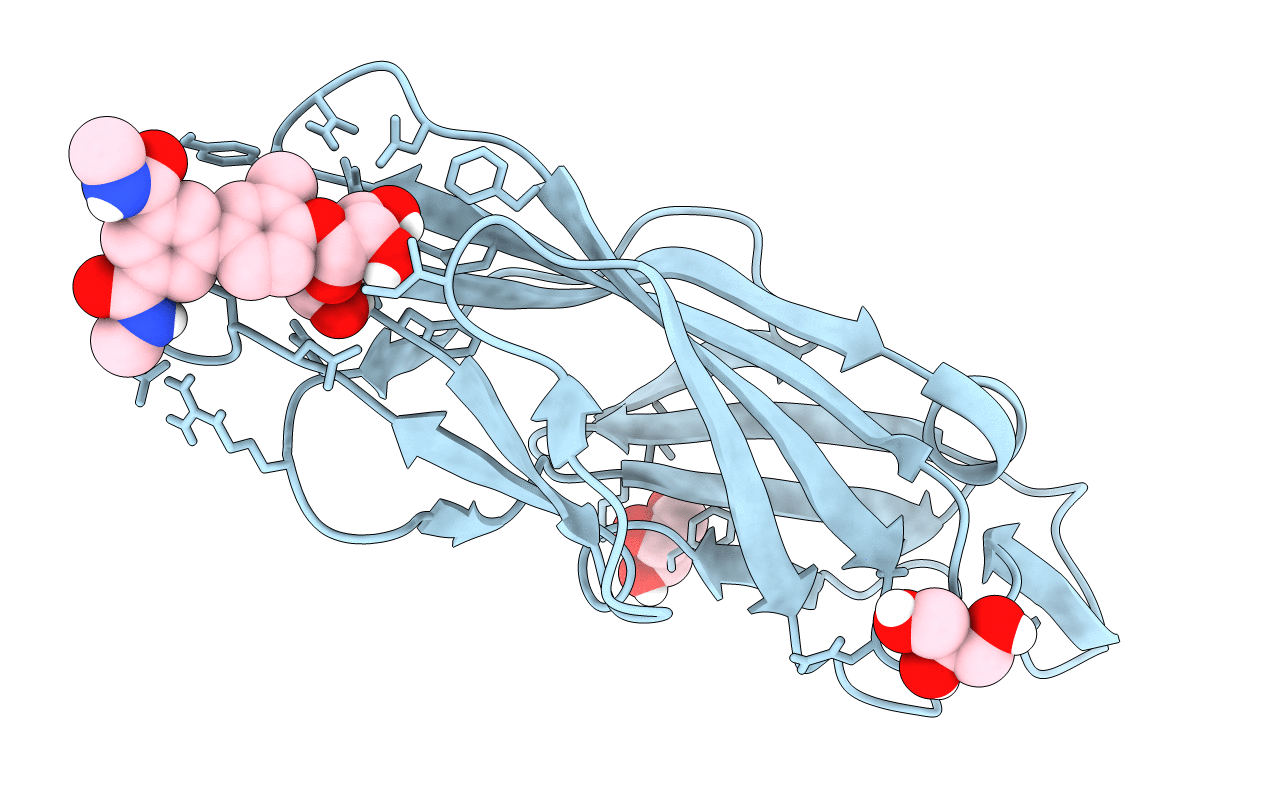
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-07-20 Classification: MEMBRANE PROTEIN Ligands: NAG, VIS, NA, MPG |
 |
Crystal Structure Of Para-Biphenyl-2-Methyl-3', 5' Di-Methyl Amide Mannoside Bound To Fimh Lectin Domain
Organism: Escherichia coli j96
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2016-05-04 Classification: SUGAR BINDING PROTEIN Ligands: 5U7, GOL |
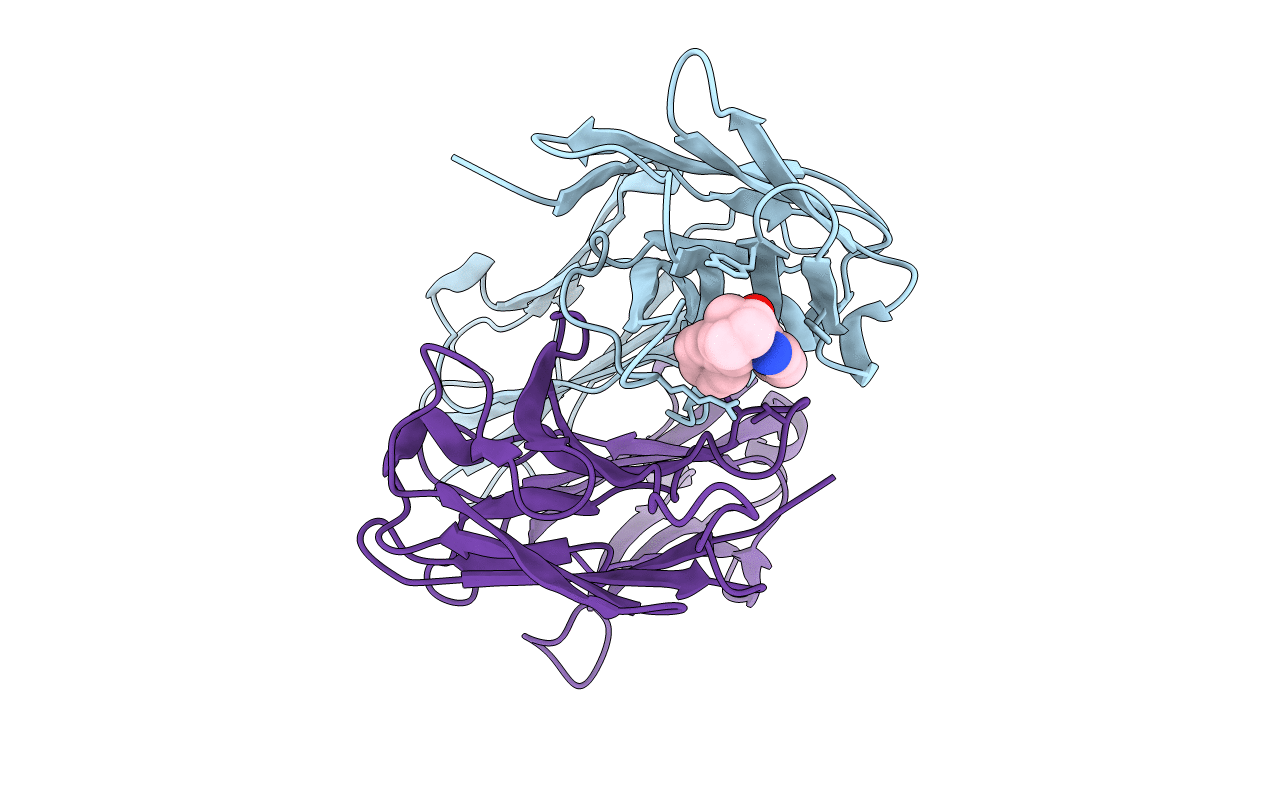 |
Structural Basis For Disfavored Elimination Reaction In Catalytic Antibody 1D4
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-12-05 Classification: IMMUNE SYSTEM Ligands: OH, HBC |
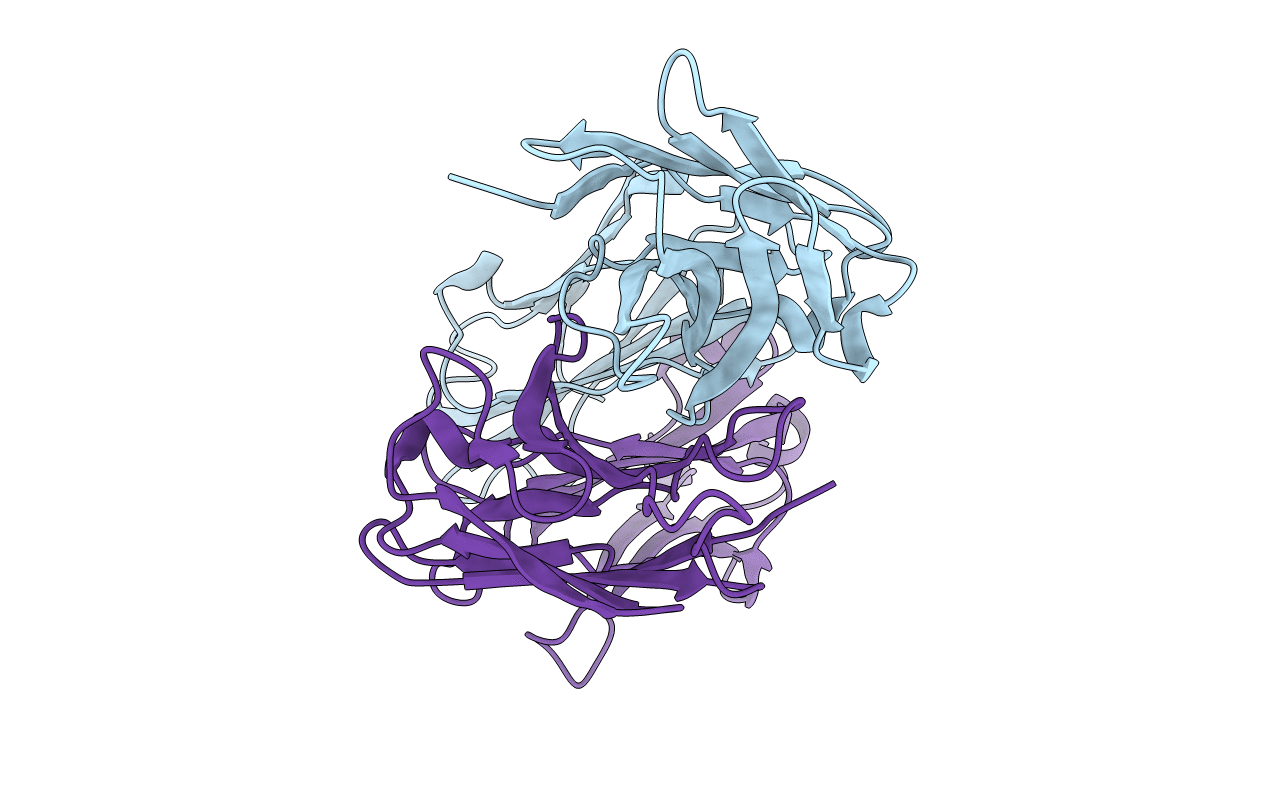 |
Structural Basis For Disfavored Elimination Reaction In Catalytic Antibody 1D4
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2001-12-05 Classification: IMMUNE SYSTEM |

