Search Count: 19
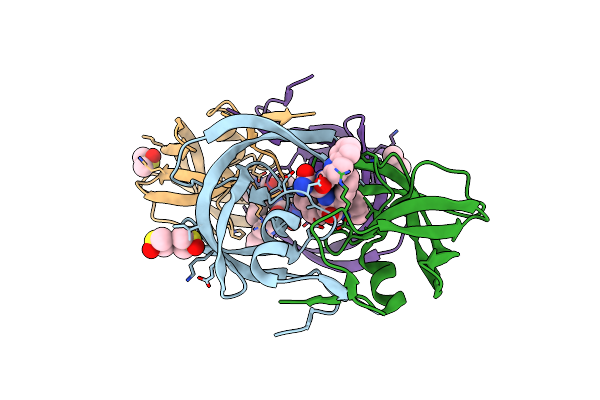 |
New Crystal Form Of Hiv-1 Protease/Saquinavir Structure Reveals Carbamylation Of N-Terminal Proline
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2010-06-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMS, ROC, CL |
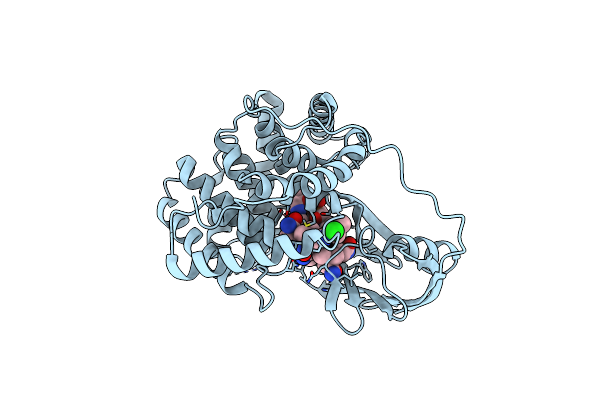 |
Crystallographic Analysis Of Upper Axial Ligand Substitutions In Cobalamin Bound To Transcobalamin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: B12, CYN, CL |
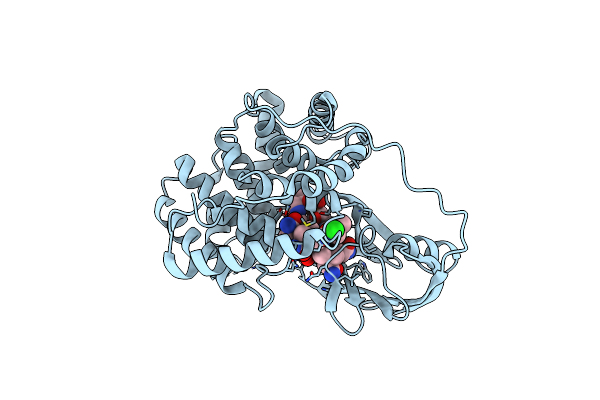 |
Crystallographic Analysis Of Beta-Axial Ligand Substitutions In Cobalamin Bound To Transcobalamin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: B12, CL, SO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: B12 |
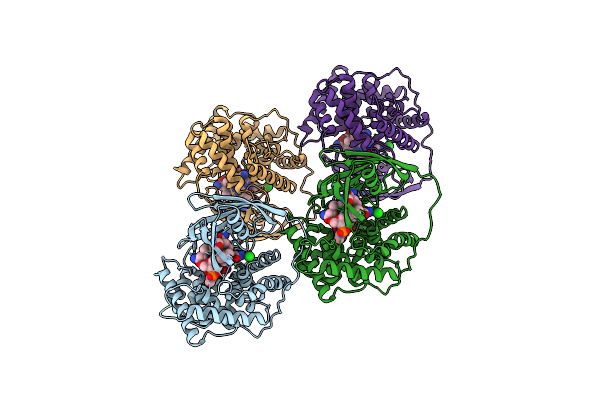 |
Structure Of Cobalamin-Complexed Bovine Transcobalamin In Monoclinic Crystal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: CL, B12 |
 |
Structure Of Cobalamin-Complexed Bovine Transcobalamin In Trigonal Crystal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: CL, B12 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-02-21 Classification: HYDROLASE Ligands: DMS, NA, CL, ACT, IPF, GOL |
 |
Method: X-RAY DIFFRACTION
Resolution:1.25 Å Release Date: 2004-12-14 Classification: METAL BINDING PROTEIN Ligands: CL, DEU |
 |
Method: X-RAY DIFFRACTION
Resolution:2.99 Å Release Date: 2004-05-18 Classification: DE NOVO PROTEIN Ligands: MN |
 |
Method: X-RAY DIFFRACTION
Resolution:3.10 Å Release Date: 2004-04-06 Classification: DE NOVO PROTEIN Ligands: CO |
 |
Method: X-RAY DIFFRACTION
Resolution:2.90 Å Release Date: 2004-04-06 Classification: DE NOVO PROTEIN Ligands: CO |
 |
Sliding Helix Induced Change Of Coordination Geometry In A Model Di-Mn(Ii) Protein
Method: X-RAY DIFFRACTION
Resolution:1.91 Å Release Date: 2003-05-20 Classification: DE NOVO PROTEIN Ligands: MN |
 |
Structure Determination Of The Ferricytochrome C2 From Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2002-01-16 Classification: ELECTRON TRANSPORT Ligands: SO4, HEC, GOL |
 |
Method: X-RAY DIFFRACTION
Resolution:1.70 Å Release Date: 2002-01-16 Classification: DE NOVO PROTEIN Ligands: MN, DMS |
 |
Method: X-RAY DIFFRACTION
Resolution:2.20 Å Release Date: 2002-01-16 Classification: DE NOVO PROTEIN Ligands: MN, DMS |
 |
Rhodopseudomonas Palustris Cyt C2 Ammonia Complex At 1.15 Angstrom Resolution
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2001-04-04 Classification: ELECTRON TRANSPORT Ligands: SO4, HEC, NH3 |
 |
Structure Determination Of The Ferrocytochrome C2 From Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2001-04-04 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Refined Crystal Structure Of Cytochrome C2 From Rhodopseudomonas Palustris At 1.4 Angstrom Resolution
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2001-01-02 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, NH3 |
 |
Method: X-RAY DIFFRACTION
Resolution:2.50 Å Release Date: 2000-07-26 Classification: DE NOVO PROTEIN Ligands: ZN |

