Search Count: 23
 |
Crystal Structure Of Kinase Domain Of Her2 Exon 20 Insertion Mutant In Complex With Tucatinib
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: A1AAC, EDO, CL, PEG |
 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With Cefepime
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-09-04 Classification: SIGNALING PROTEIN Ligands: UJ9 |
 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With A Boronate Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-10 Classification: SIGNALING PROTEIN Ligands: SZI |
 |
Crystal Structure Of S. Aureus Blar1 Sensor Domain In Complex With An Imidazole Inhibitor
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-10 Classification: SIGNALING PROTEIN Ligands: SYU |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: SO4, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx From P. Aeruginosa In Complex With 1-Deoxynojirimycin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, NOJ, MG |
 |
The Crystal Structure The Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Two Glucose Molecules
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Glucose
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: BGC, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Cellobiose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, PGE, MG |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Lactose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PEG, MG, PGE |
 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant From P. Aeruginosa In Complex With Laminaritriose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: MG |
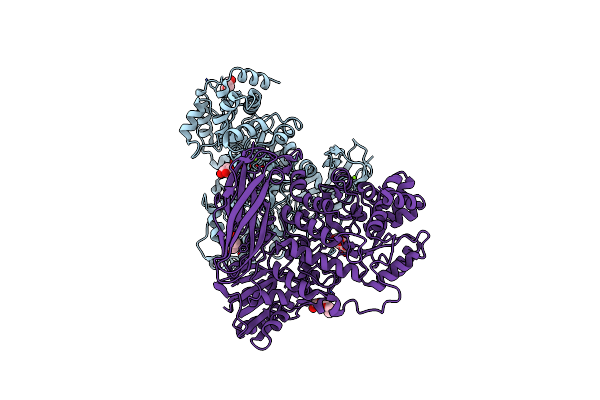 |
The Crystal Structure Of Glycoside Hydrolase Bglx Inactive Mutant D286N From P. Aeruginosa In Complex With Xylotriose
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, PEG, MG |
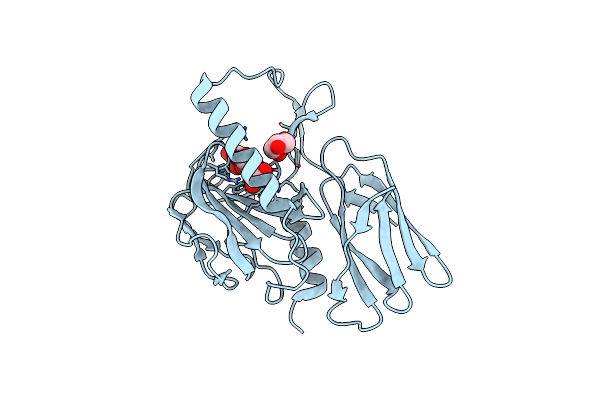 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN, TAR, GOL |
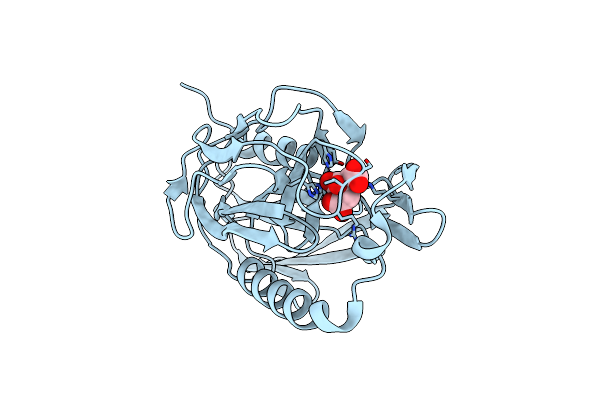 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN, CIT |
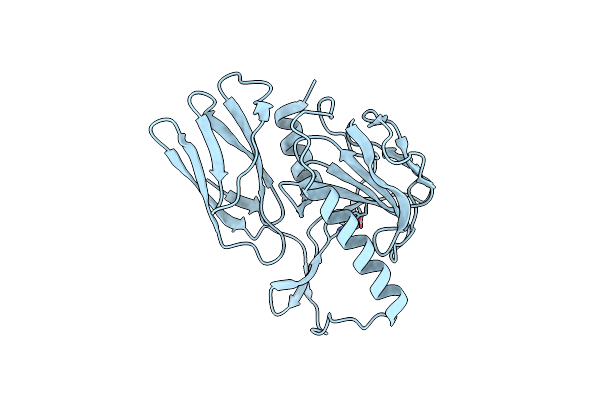 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of H247A Mutant Peptidoglycan Peptidase Complex With Tetra-Tri Peptide
Organism: Campylobacter jejuni, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of H247A Mutant Peptidoglycan Peptidase Complex With Penta Peptide
Organism: Campylobacter jejuni, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN, TAR |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-01-15 Classification: HYDROLASE Ligands: ZN, CIT |

