Search Count: 10
 |
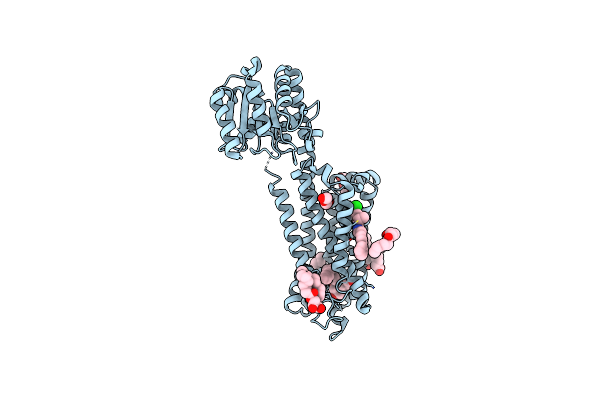
Structural Mechanism Of Cb1R Binding To Peripheral And Biased Inverse Agonists
Organism: Homo sapiens, Pyrococcus abyssi ge5, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: 7DY |
 |
Structural Mechanism Of Cb1R Binding To Peripheral And Biased Inverse Agonists
Organism: Lama glama, Homo sapiens, Pyrococcus abyssi ge5
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: A1AKO |
 |
Structural Mechanism Of Cb1R Binding To Peripheral And Biased Inverse Agonists
Organism: Homo sapiens, Pyrococcus abyssi ge5, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: A1AKN |
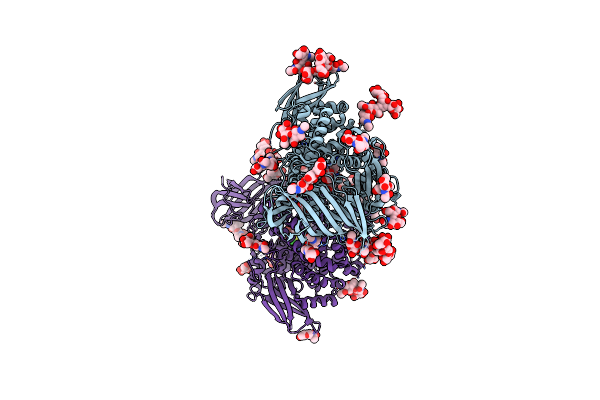 |
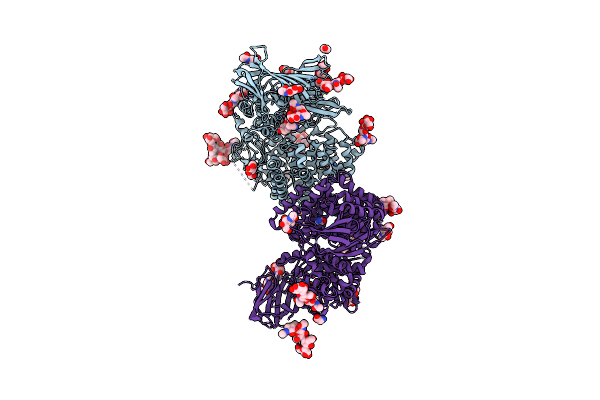
Insulin Regulated Aminopeptidase (Irap) In Complex With An Allosteric Aryl Sulfonamide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: NAG, MLT, ZN, UKG, PEG, EDO |
 |
Insulin-Regulated Aminopeptidase (Irap) In Complex With An Allosteric Benzopyran-Based Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2024-01-31 Classification: HYDROLASE Ligands: ZN, UJX, NAG, PEG, EDO, GLY |
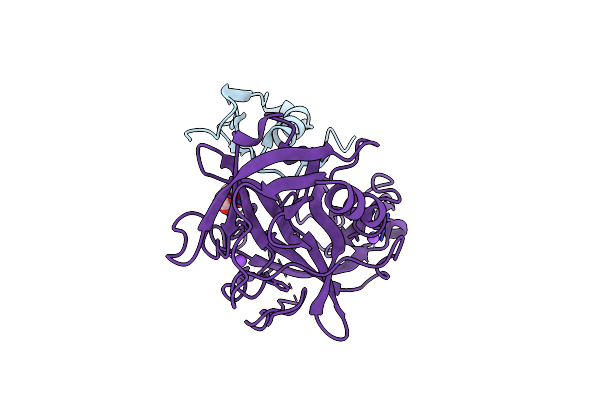 |
Crystal Structure Of Human Microplasmin In Complex With Kazal-Type Inhibitor Aati
Organism: Aedes aegypti, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: GOL, NA |
 |
Organism: Homo sapiens, Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2019-10-23 Classification: CELL CYCLE Ligands: 9GF, 9GL, OLA, OLC, PEG |
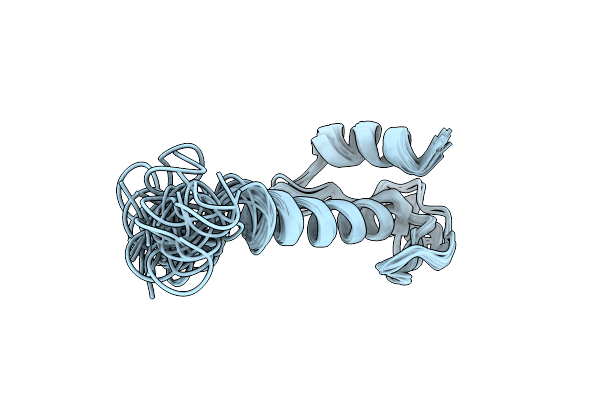 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-08-08 Classification: HYDROLASE INHIBITOR |
 |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-10-14 Classification: Membrane protein/Chaperone Ligands: MG |

