Search Count: 16
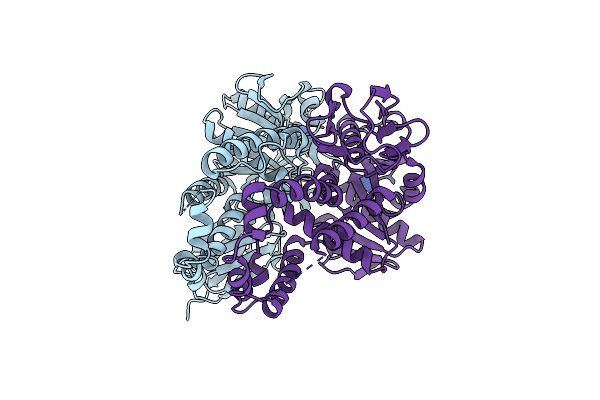 |
Re-Refinement Of Crystal Structure Of Nosget3D, The All4481 Protein From Nostoc Sp. Pcc 7120
Organism: Nostoc sp. pcc 7120 = fachb-418
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-05-24 Classification: CHAPERONE Ligands: AZI |
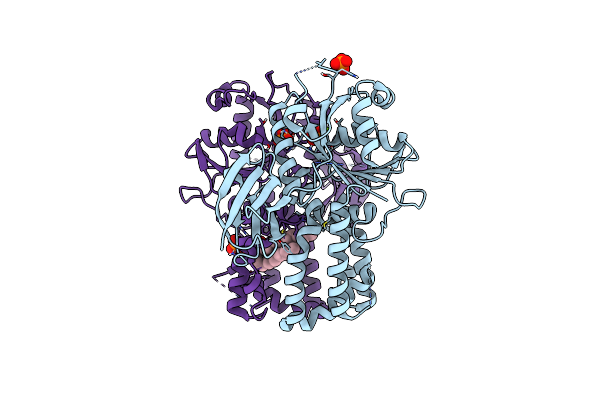 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-03 Classification: CHAPERONE Ligands: PO4, MG, XP4 |
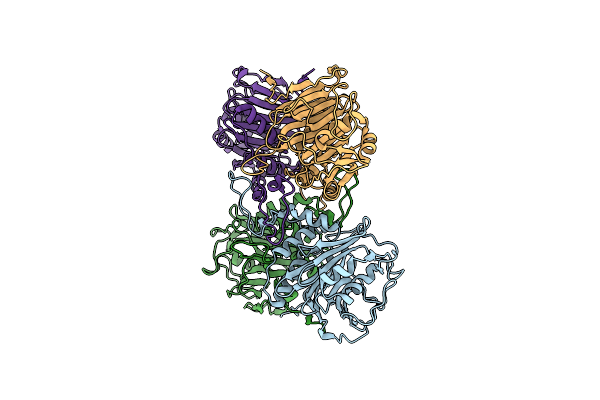 |
Organism: Enterococcus faecalis t2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-10 Classification: HYDROLASE |
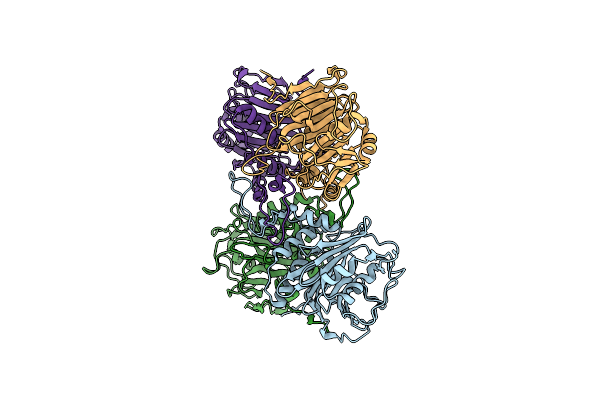 |
Organism: Enterococcus faecalis t2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-10 Classification: HYDROLASE |
 |
Organism: Cicer arietinum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-06-20 Classification: HYDROLASE INHIBITOR |
 |
Organism: Shewanella loihica pv-4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-03-07 Classification: HYDROLASE |
 |
Unique Choloylglycine Hydrolase(Cgh) Member Mutant (C1S) From Shewanella Loihica Pv-4
Organism: Shewanella loihica pv-4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: OOA, GOL |
 |
Crystal Structure Determination Of Cys2Ala Mutant Of Bile Salt Hydrolase From Enterococcus Feacalis
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-16 Classification: HYDROLASE |
 |
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-06-21 Classification: HYDROLASE |
 |
Crystal Structure Determination Of Bile Salt Hydrolase From Enterococcus Feacalis
Organism: Enterococcus faecalis t2
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-11-11 Classification: HYDROLASE |
 |
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-10-07 Classification: HYDROLASE |
 |
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-07-22 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of S1G Mutant Of Penicillin G Acylase From Kluyvera Citrophila
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-07-22 Classification: HYDROLASE Ligands: CA |
 |
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2013-05-01 Classification: PROTEIN TRANSPORT |
 |
Organism: Aquifex aeolicus vf5, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:6.80 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-04-28 Classification: CHAPERONE |

