Search Count: 21
 |
Hiv-1 Reverse Transcriptase With New Non-Nucleoside Reverse Transcriptase Inhibitor 12126065
Organism: Hiv-1 06tg.ht008
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRAL PROTEIN Ligands: A1BYY |
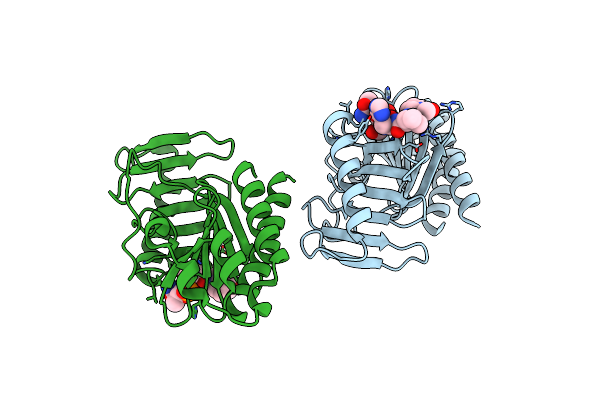 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-09-25 Classification: STRUCTURAL PROTEIN |
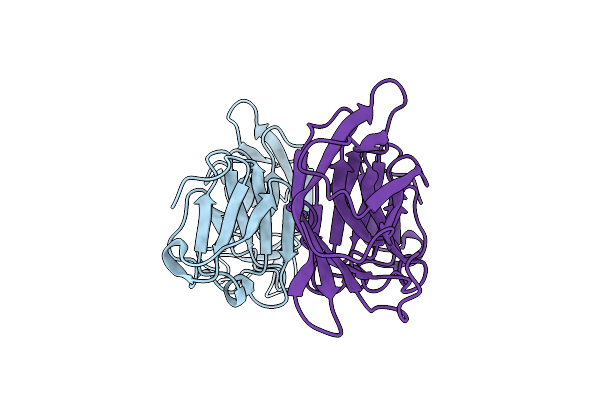 |
Crystal Structure Of Abundant Perithecial Protein (App) From Neurospora Crassa
Organism: Neurospora crassa (strain atcc 24698 / 74-or23-1a / cbs 708.71 / dsm 1257 / fgsc 987)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-01-23 Classification: METAL BINDING PROTEIN |
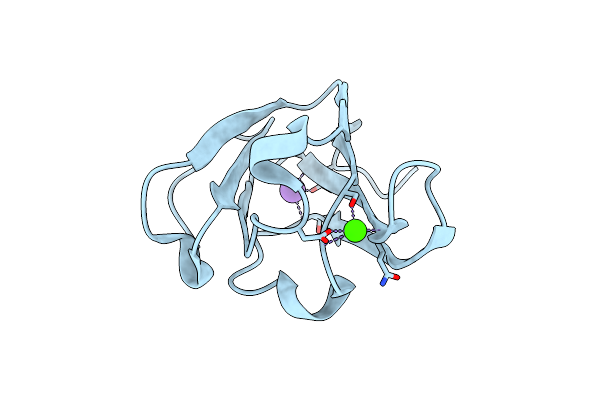 |
Crystal Structure Of A Beta Gamma-Crystallin Domain Of Abundant Perithecial Protein (App) From Neurospora Crassa In The Ca2+-Bound Form
Organism: Neurospora crassa (strain atcc 24698 / 74-or23-1a / cbs 708.71 / dsm 1257 / fgsc 987)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-01-23 Classification: METAL BINDING PROTEIN Ligands: CA, K |
 |
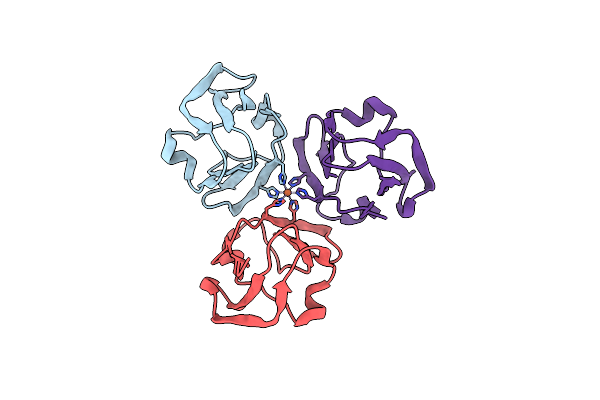
Crystal Structure Of The Ntd_N/C Domain Of Alkylhydroperoxide Reductase Ahpf From Enterococcus Faecalis (V583)
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, TRS, PRO |
 |
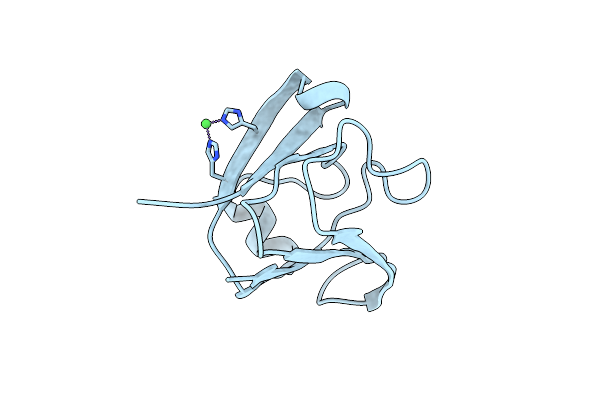
Crystal Structure Of A Transition-Metal-Ion-Binding Betagamma-Crystallin From Methanosaeta Thermophila
Organism: Methanosaeta thermophila pt
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-02-01 Classification: METAL BINDING PROTEIN Ligands: FE2 |
 |
Crystal Structure Of Clostrillin Double Mutant (S17H,S19H) In Complex With Nickel
Organism: Clostridium beijerinckii ncimb 8052
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-02-01 Classification: METAL BINDING PROTEIN Ligands: NI |
 |
Crystal Structure Of H5 Hemagglutinin Q226L Mutant From The Influenza Virus A/Duck/Egypt/10185Ss/2010 (H5N1)
Organism: Influenza a virus (a/duck/egypt/10185ss/2010(h5n1)), Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-12-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of H5 Hemagglutinin Q226L Mutant From The Influenza Virus A/Duck/Egypt/10185Ss/2010 (H5N1) With Lsta
Organism: Influenza a virus (a/duck/egypt/10185ss/2010(h5n1)), Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2015-12-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of H5 Hemagglutinin Q226L Mutant From The Influenza Virus A/Duck/Egypt/10185Ss/2010 (H5N1) With Lstc
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of H5 Hemagglutinin Mutant (N224K, Q226L, N158D And L133A Deletion) From The Influenza Virus A/Chicken/Vietnam/Ncvd-093/2008 (H5N1)
Organism: Influenza a virus (a/chicken/vietnam/ncvd-093/2008(h5n1)), Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of H5 Hemagglutinin Mutant (N224K, Q226L, N158D And L133A Deletion) From The Influenza Virus A/Chicken/Vietnam/Ncvd-093/2008 (H5N1) With Lsta
Organism: Influenza a virus (a/chicken/vietnam/ncvd-093/2008(h5n1)), Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of H5 Hemagglutinin Mutant (N224K, Q226L, N158D And L133A Deletion) From The Influenza Virus A/Chicken/Vietnam/Ncvd-093/2008 (H5N1) With Lstc
Organism: Influenza a virus (a/chicken/vietnam/ncvd-093/2008(h5n1)), Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Solution Structure Of The Terminal Ig-Like Domain From Leptospira Interrogans Ligb
Organism: Leptospira interrogans
Method: SOLUTION NMR Release Date: 2014-08-13 Classification: CELL ADHESION |
 |
Directed Evolution And Rational Design Of A De Novo Designed Esterase Toward Improved Catalysis. Northeast Structural Genomics Consortium (Nesg) Target Or184
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2013-05-22 Classification: Structural Genomics, Unknown Function Ligands: BR, GOL, PE5 |
 |
X-Ray Structure Of The C-Terminal Domain (277-440) Of Putative Chitobiase From Bacteroides Thetaiotaomicron. Northeast Structural Genomics Consortium Target Btr324A.
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-03-31 Classification: structural genomics, unknown function Ligands: ZN, PEG |
 |
Crystal Structure Of Putative Thioesterase Yhda From Lactococcus Lactis. Northeast Structural Genomics Consortium Target Kr113
Organism: Lactococcus lactis subsp. lactis
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2009-03-17 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Crystal Structure Of N-Terminal Actin-Binding Domain From Human Filamin B (Tandem Ch-Domains). Northeast Structural Genomics Consortium Target Hr5571A.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-01-06 Classification: actin binding protein Ligands: ACY |
 |
Crystal Structure Of Chondroitinase B Complexed To Dermatan Sulfate Hexasaccharide
Organism: Pedobacter heparinus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-06-10 Classification: LYASE Ligands: CA |
 |
Crystal Structure Of Chondroitinase B Complexed To Chondroitin 4-Sulfate Tetrasaccharide
Organism: Pedobacter heparinus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-04-19 Classification: LYASE |

