Search Count: 67
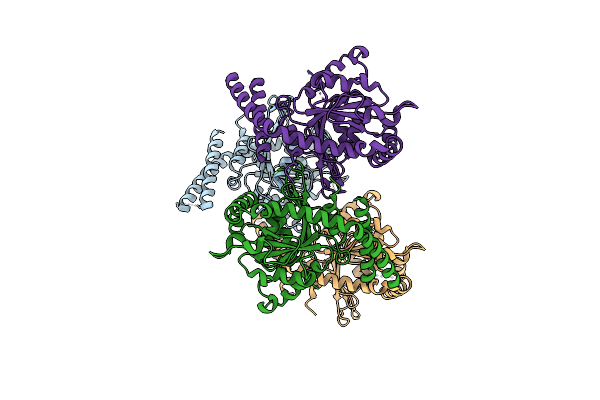 |
Crystal Structure Of Ct-Slim-Deleted Human Drp1 Gtpase Domain-Bse Fusion Protein In The Apo State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: MOTOR PROTEIN |
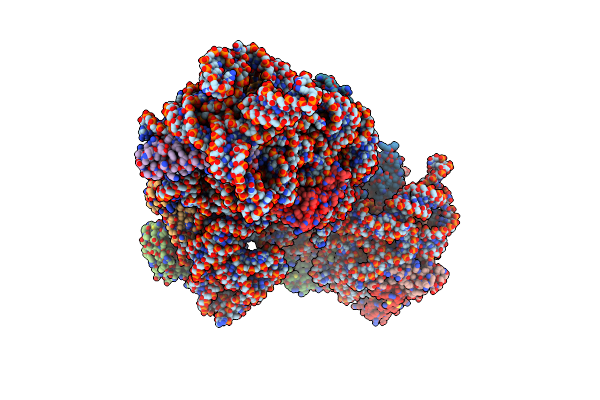 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: RIBOSOME |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: RIBOSOME |
 |
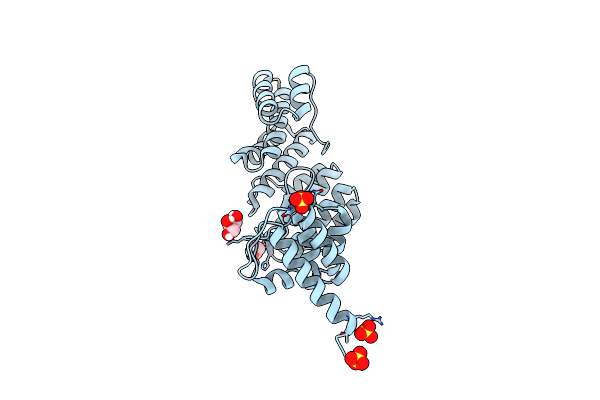
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: CYTOSOLIC PROTEIN |
 |
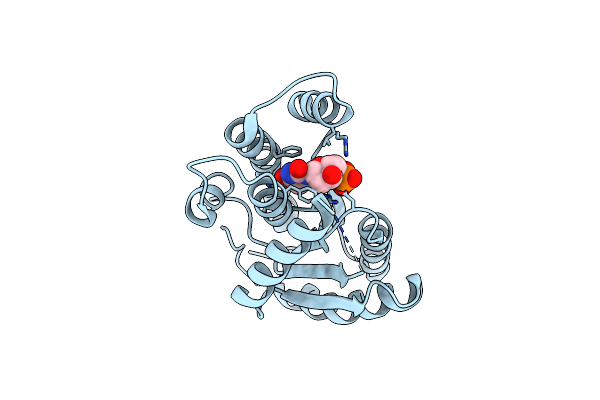
Crystal Structure And Biophysical Characterization Of Tpr Domain Of Ecca5 From Esx-5 Pathway Of Mycobacterium Tuberculosis H37Rvr
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-09-14 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of Thymidylate Kinase With Tmp And Its Low-Resolution (Saxs) Solution Structure From Brugia Malayi
Organism: Brugia malayi
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-07-27 Classification: TRANSFERASE Ligands: TMP |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-02-10 Classification: LIGASE Ligands: GOL, EWO, SO4 |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-08-26 Classification: LIGASE Ligands: AMP, NMN, SO4 |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-08-26 Classification: LIGASE Ligands: AMP, SO4 |
 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-08-26 Classification: LIGASE Ligands: NMN, SO4 |
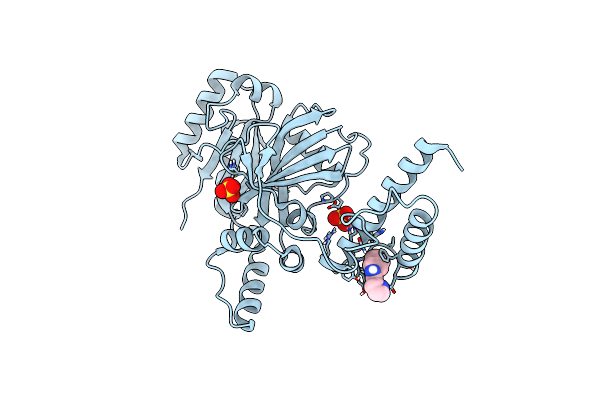 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2020-07-29 Classification: LIGASE Ligands: DKR, SO4 |
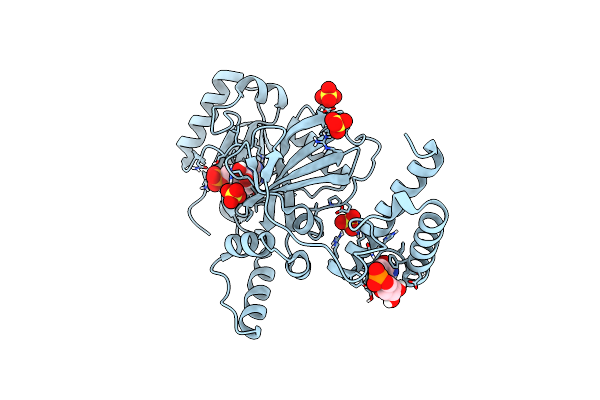 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-22 Classification: LIGASE Ligands: NMN, AMP, SO4 |
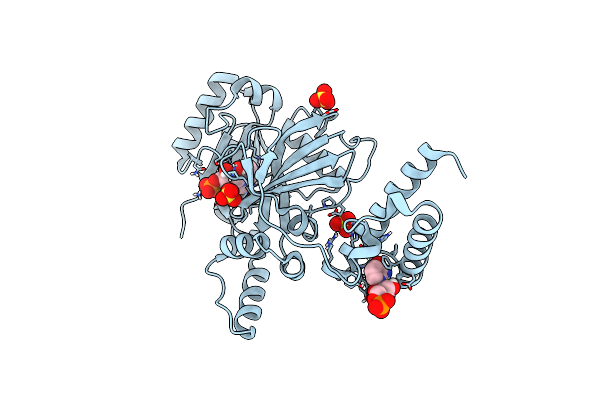 |
Structural Basis For Domain Rotation During Adenylation Of Active Site K123 And Fragment Library Screening Against Nad+ -Dependent Dna Ligase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-01 Classification: DNA BINDING PROTEIN Ligands: AMP, NMN, SO4 |
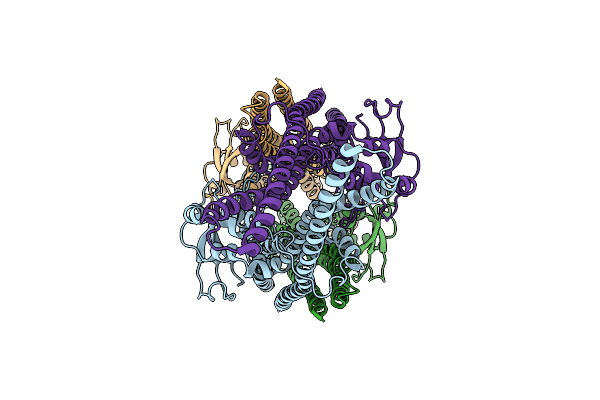 |
Organism: Saccharomyces cerevisiae w303
Method: ELECTRON MICROSCOPY Release Date: 2020-04-01 Classification: LIPID BINDING PROTEIN |
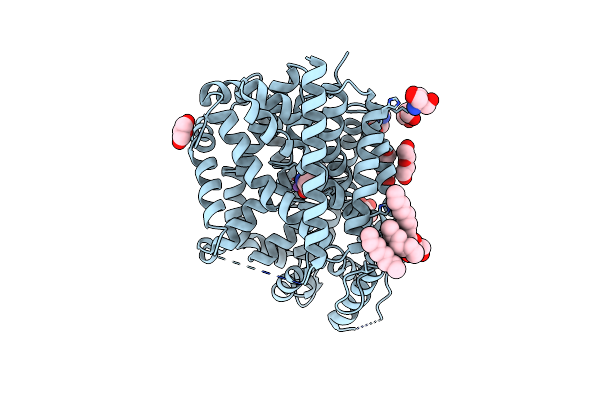 |
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2020-01-15 Classification: MEMBRANE PROTEIN Ligands: LEU, NA, GOL, PEG, TRS, BOG, OCT |
 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-06-05 Classification: IMMUNE SYSTEM |
 |
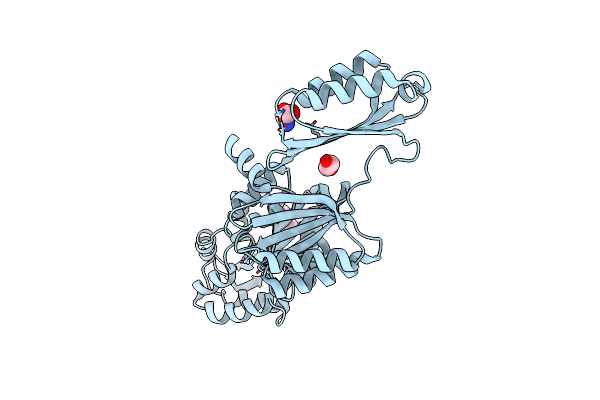
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-06-05 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Mycobacterium Avium Serb2 In Complex With Serine At Act Domain
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: EDO, PG0, MG, SER |
 |
Crystal Structure Of Mycobacterium Avium Serb2 With Serine Present At Slightly Different Position Near Act Domain
Organism: Mycobacterium avium (strain 104)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: MG, EDO, PG0, SER |

