Search Count: 19
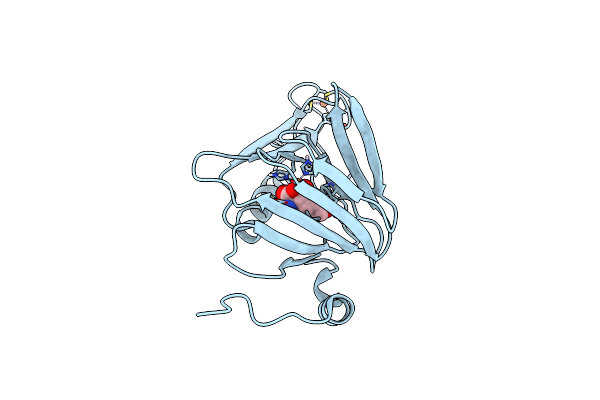 |
Understanding Extradiol Dioxygenase Mechanism In Nad+ Biosynthesis By Viewing Catalytic Intermediates - 2,3-Cis-4,5-Trans Acms Bound To I142A Mutant Hao
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2016-02-10 Classification: OXIDOREDUCTASE Ligands: FE2, 1UC |
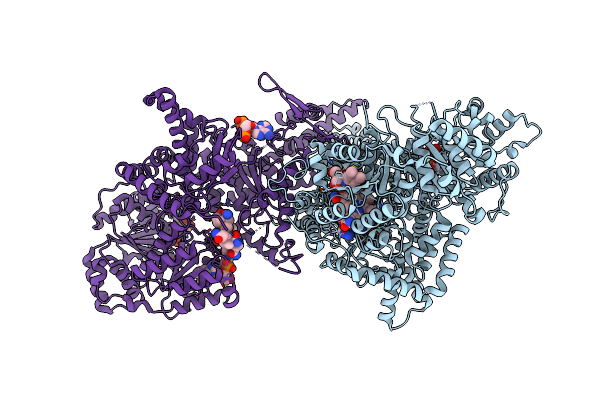 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Isobutyryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, CO6, GDP, MG |
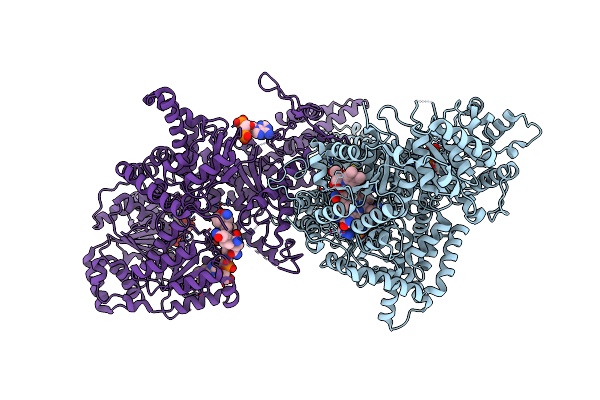 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate N-Butyryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, BCO, GDP, MG |
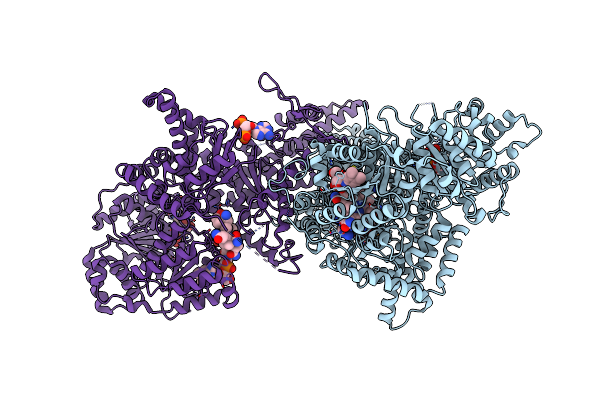 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Isovaleryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, IVC, GDP, MG |
 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Pivalyl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, 52O, GDP, MG |
 |
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2015-07-08 Classification: HYDROLASE Ligands: CD, TCE |
 |
High-Resolution Structure Of The Ni-Bound Form Of The Y135F Mutant Of C. Metallidurans Cnrxs
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2015-02-11 Classification: METAL BINDING PROTEIN Ligands: NI, CL, CO2, K, FMT |
 |
High-Resolution Structure Of The Co-Bound Form Of The Y135F Mutant Of C. Metallidurans Cnrxs
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-02-11 Classification: METAL BINDING PROTEIN Ligands: CO, CL, CO2, K, FMT |
 |
High-Resolution Structure Of Two Ni-Bound Forms Of The M123C Mutant Of C. Metallidurans Cnrxs
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-02-11 Classification: METAL BINDING PROTEIN Ligands: NI, NA |
 |
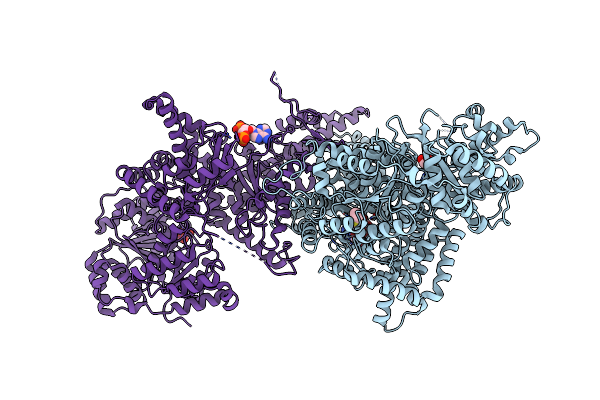
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, And Mg (Holo-Icmf/Gdp)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: B12, 5AD, GDP, MG, ACT |
 |
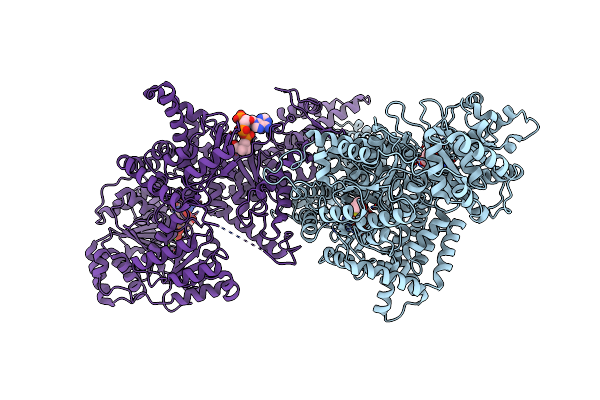
Isobutyryl-Coa Mutase Fused With Bound Butyryl-Coa And Without Cobalamin Or Gdp (Apo-Icmf)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: BCO, TLA |
 |
Isobutyryl-Coa Mutase Fused With Bound Butyryl-Coa, Gdp, And Mg And Without Cobalamin (Apo-Icmf/Gdp)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: BCO, GDP, MG |
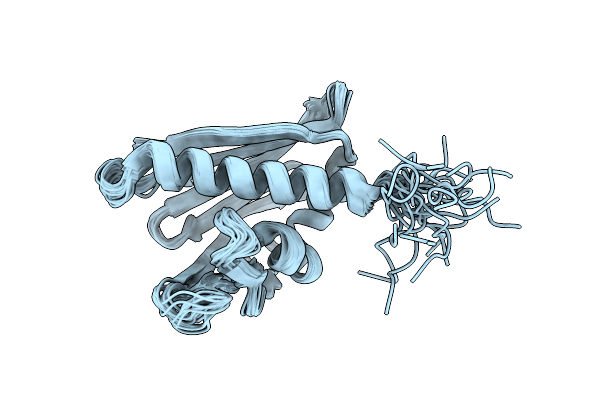 |
Solution Nmr Structure Of Protein Rmet_5065 From Ralstonia Metallidurans, Northeast Structural Genomics Consortium Target Crr115
Organism: Ralstonia metallidurans
Method: SOLUTION NMR Release Date: 2011-05-18 Classification: Structural genomics, Unknown function |
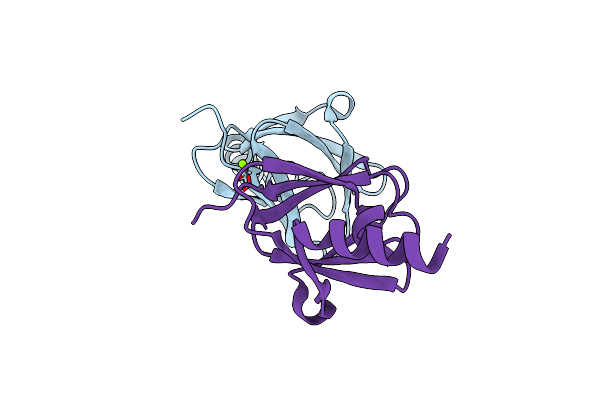 |
Organism: Ralstonia metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-08-18 Classification: METAL BINDING PROTEIN Ligands: CL, MG |
 |
Organism: Ralstonia metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-08-18 Classification: METAL BINDING PROTEIN Ligands: PEG, EDO, CU, MG, CL, TRS |
 |
Organism: Ralstonia metallidurans
Method: SOLUTION NMR Release Date: 2010-03-16 Classification: METAL BINDING PROTEIN Ligands: CU1 |
 |
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-03-10 Classification: METAL BINDING PROTEIN Ligands: CU1, SCN |
 |
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-10 Classification: METAL BINDING PROTEIN |
 |
X-Ray Structure Of The Metal-Sensor Cnrx In Both The Apo- And Copper-Bound Forms
Organism: Ralstonia metallidurans
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2008-11-25 Classification: METAL BINDING PROTEIN Ligands: CU |

