Search Count: 9
 |
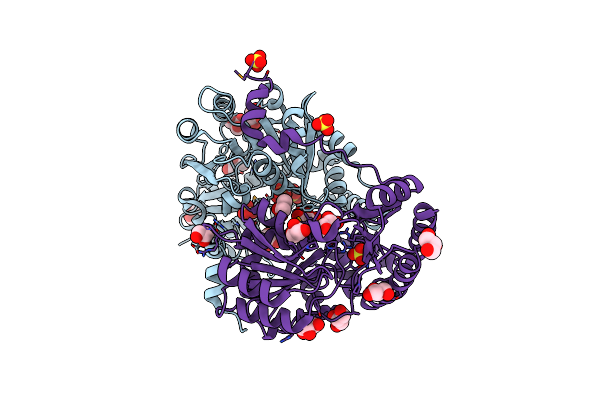
Crystal Structure Of A Putative Cysteine Dioxygnase From Ralstonia Eutropha: An Alternative Modeling Of 2Gm6 From Jcsg Target 361076
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: FE, OXY, SO4, EDO |
 |
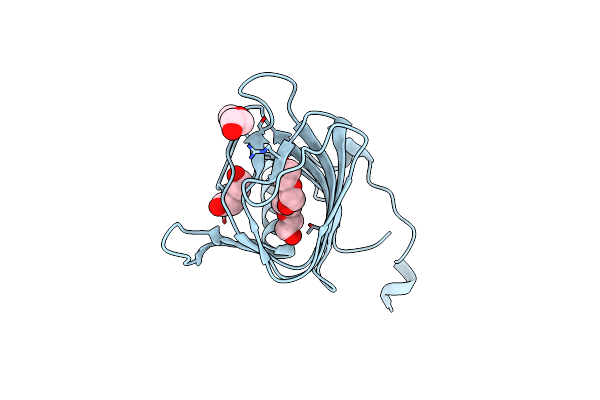
Crystal Structure Of A Putative Biosynthetic Protein With Rmlc-Like Cupin Fold (Reut_B4087) From Ralstonia Eutropha Jmp134 At 1.90 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-01-13 Classification: BIOSYNTHETIC PROTEIN |
 |
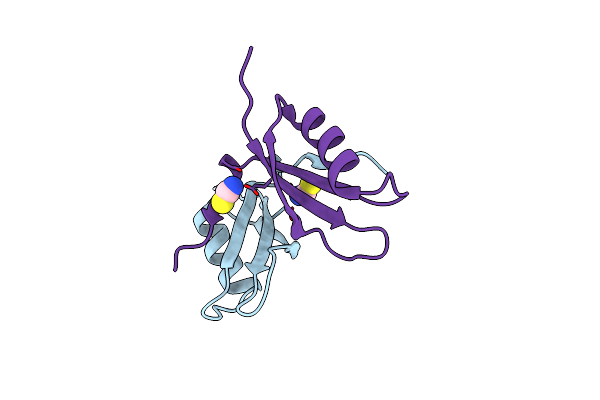
Crystal Structure Of Histidinol-Phosphate Aminotransferase (Yp_297314.1) From Ralstonia Eutropha Jmp134 At 2.05 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-11-11 Classification: TRANSFERASE Ligands: SO4, GOL |
 |
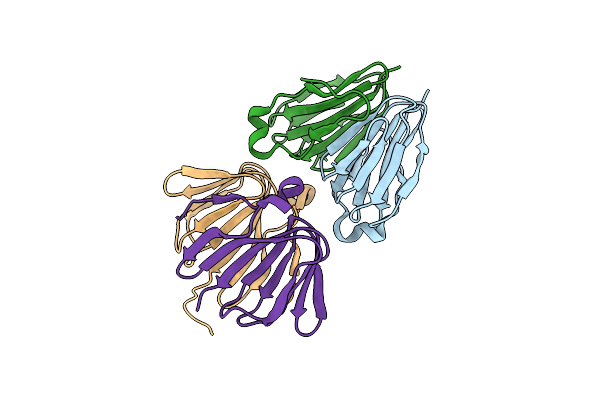
Crystal Structure Of A Phenylacetate-Coa Oxygenase Subunit Paab (Reut_A2307) From Ralstonia Eutropha Jmp134 At 2.65 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2008-09-30 Classification: OXIDOREDUCTASE Ligands: SCN |
 |
Crystal Structure Of An Rmlc-Like Cupin Protein (Reut_A0381) From Ralstonia Eutropha Jmp134 At 2.60 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-09-09 Classification: UNKNOWN FUNCTION Ligands: PEG |
 |
Crystal Structure Of A Putative Aminoglycoside Phosphotransferase (Reut_A1007) From Ralstonia Eutropha Jmp134 At 2.32 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: CA, ACT, EDO |
 |
Crystal Structure Of An Ethd-Like Protein (Reut_B5694) From Ralstonia Eutropha Jmp134 At 2.10 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-12-04 Classification: UNKNOWN FUNCTION Ligands: EDO, IPA |
 |
Crystal Structure Of Gcn5-Related N-Acetyltransferase (Yp_295895.1) From Ralstonia Eutropha Jmp134 At 1.80 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-06-05 Classification: TRANSFERASE Ligands: ZN, MG, UNL, EDO, FMT |
 |
Crystal Structure Of A Putative Thioesterase, Phenylacetic Acid Degradation-Related Protein (Reut_B4779) From Ralstonia Eutropha Jmp134 At 2.20 A Resolution
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-05-01 Classification: HYDROLASE |

