Search Count: 57
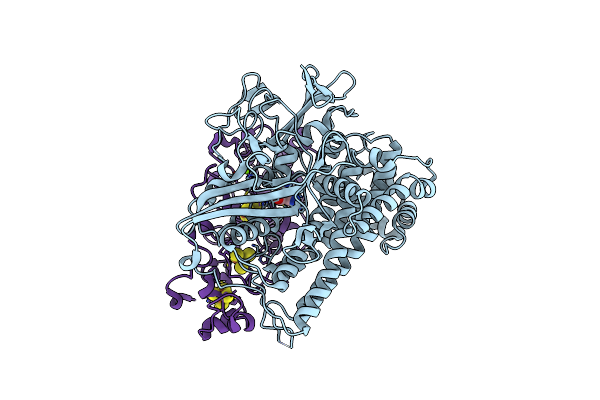 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In A Its As-Isolated High-Pressurized Form
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, F4S, CL |
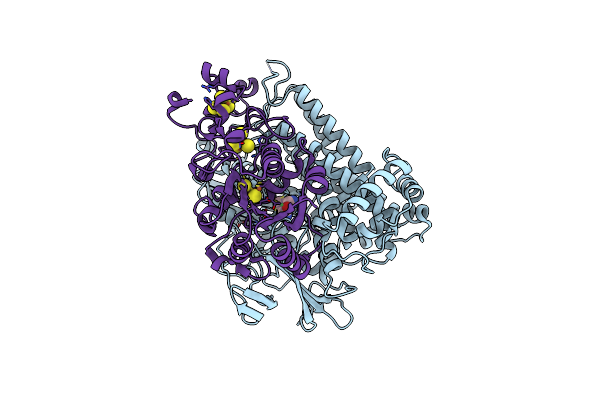 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form (Oxidized State - State 3)
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: MG, CL, NFV, SF4, F3S, F4S |
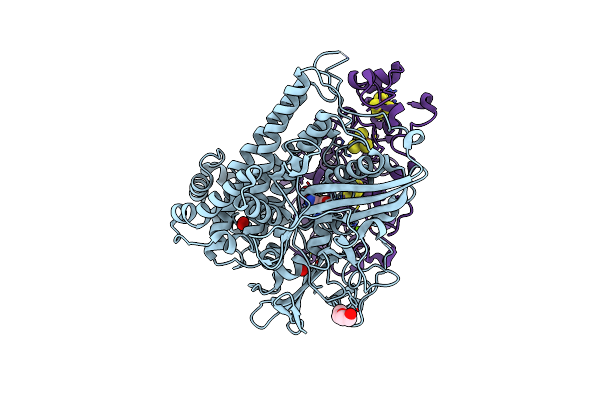 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its O2-Derivatized Form By A "Soak-And-Freeze" Derivatization Method
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-02-21 Classification: OXIDOREDUCTASE Ligands: NFV, MG, OXY, PEG, SF4, F3S, F4S, CL, O |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase Paah1 In Complex With Acetoacetyl-Coa
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-04-22 Classification: OXIDOREDUCTASE Ligands: CAA |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase Paah1 From Ralstonia Eutropha
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-02-11 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of (S)-3-Hydroxybutyryl-Coa Dehydrogenase Paah1 In Complex With Nad+
Organism: Ralstonia eutropha h16
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2015-02-11 Classification: OXIDOREDUCTASE Ligands: NAD |
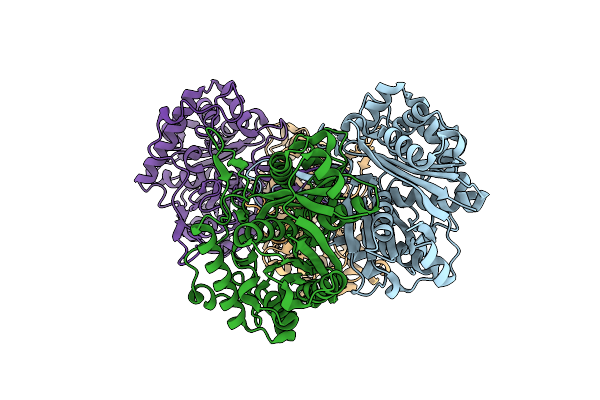 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-02-04 Classification: TRANSFERASE |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form - Oxidized State 3
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-01-28 Classification: OXIDOREDUCTASE Ligands: NFV, PO4, MG, CL, NA, SF4, F3S, F4S |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2014-12-17 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-12-17 Classification: TRANSFERASE Ligands: COA |
 |
Crystal Structure Of A Putative Cysteine Dioxygnase From Ralstonia Eutropha: An Alternative Modeling Of 2Gm6 From Jcsg Target 361076
Organism: Ralstonia eutropha jmp134
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: FE, OXY, SO4, EDO |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-11-19 Classification: TRANSFERASE |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form - Oxidized State 1
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: NFV, MG, PO4, SF4, F3S, S3F, CL |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form - Oxidized State 2
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: NFV, MG, CL, SF4, F3S, S3F |
 |
Crystal Structure Of An O2-Tolerant [Nife]-Hydrogenase From Ralstonia Eutropha In Its As-Isolated Form With Ascorbate - Partly Reduced State
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, S3F, CL |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-02-26 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-02-26 Classification: OXIDOREDUCTASE Ligands: NAD, THR |
 |
Crystal Structure Of (R)-3-Hydroxybutyryl-Coa Dehydrogenase From Ralstonia Eutropha
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-12-11 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of (R)-3-Hydroxybutyryl-Coa Dehydrogenase From Ralstonia Eutropha In Complexed With Acetoacetyl-Coa
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2013-12-11 Classification: OXIDOREDUCTASE Ligands: CAA, GOL |
 |
Crystal Structure Of (R)-3-Hydroxybutyryl-Coa Dehydrogenase From Ralstonia Eutropha In Complexed With Nadp
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-11 Classification: OXIDOREDUCTASE Ligands: NAP |

