Search Count: 1,207
 |
Organism: Ralstonia solanacearum uw551
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: IMMUNOSUPPRESSANT |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG, CL |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: A1IXG, ACT, BDF, ZN |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG, ZN |
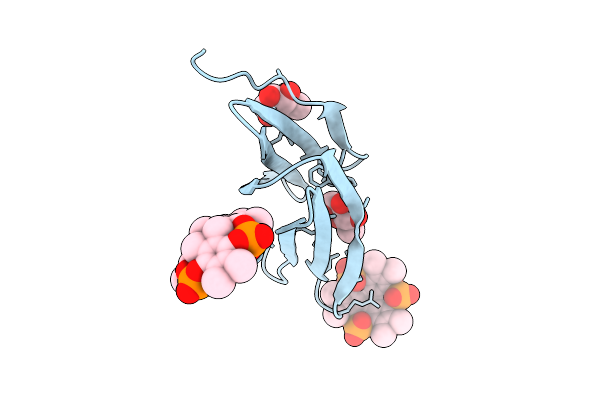 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG |
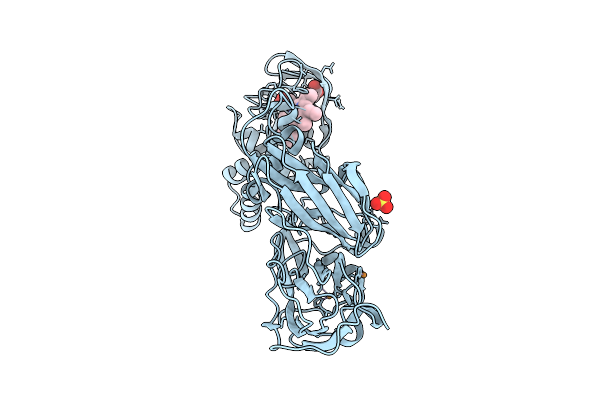 |
Crystal Structure Of Reduced Wild Type Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC, SO4 |
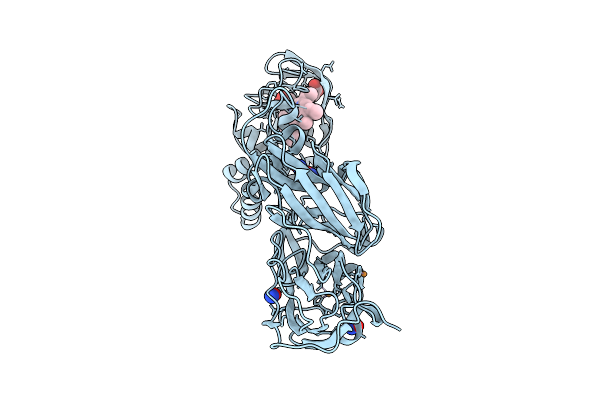 |
Crystal Structure Of Nitric Oxide Treated F295L Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC, NO |
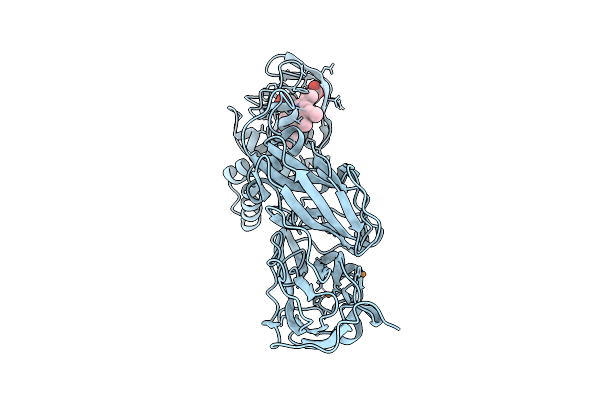 |
Crystal Structure Of Reduced F295L Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC |
 |
Crystal Structure Of Nitric Oxide-Treated Q262N Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: EVB, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: A1IZO, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IZO |
 |
Crystal Structure Of As-Isolated F295L Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: CU, HEC |
 |
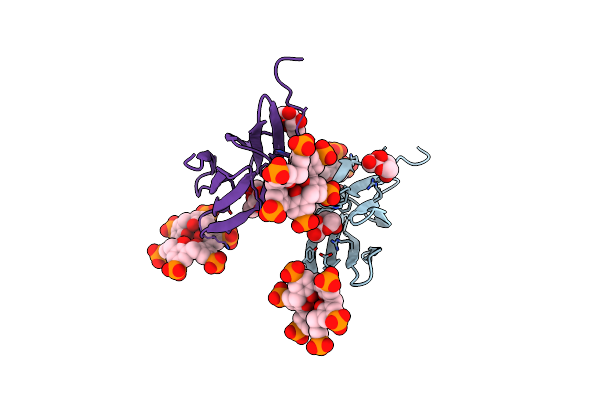
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: 7AZ, BDF |
 |
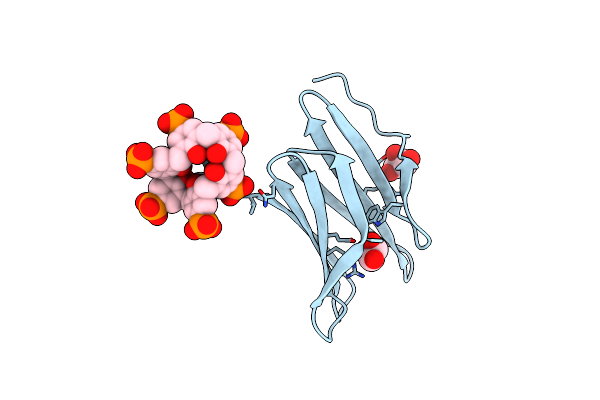
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: BDF, 7AZ, GOL |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: 7AZ, GOL |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2025-04-16 Classification: SUGAR BINDING PROTEIN Ligands: BDF, 7AZ |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: T3Y, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: BDF, T3Y |

