Search Count: 15
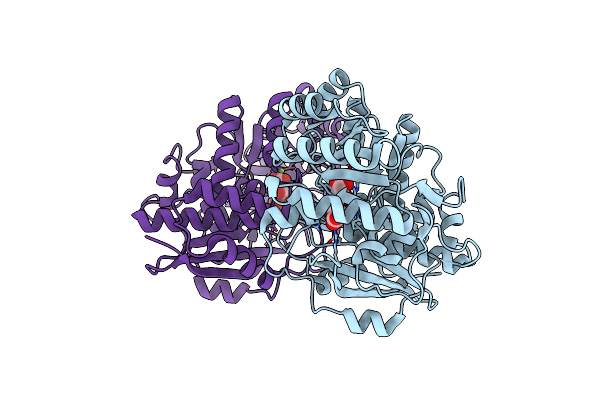 |
Crystal Structure Of The Mutant Y90F Of Divergent Galactarate Dehydratase From Oceanobacillus Iheyensis Complexed With Mg And Galactarate
Organism: Oceanobacillus iheyensis hte831
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-12-22 Classification: ISOMERASE Ligands: GAE, MG |
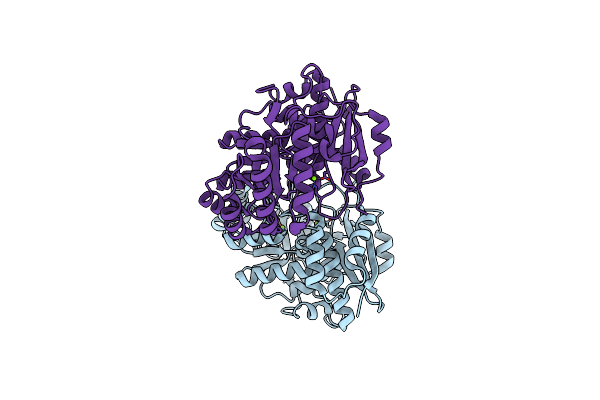 |
Crystal Structure Of Divergent Enolase From Oceanobacillus Iheyensis Complexed With Mg
Organism: Oceanobacillus iheyensis hte831
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-02-03 Classification: ISOMERASE Ligands: MG |
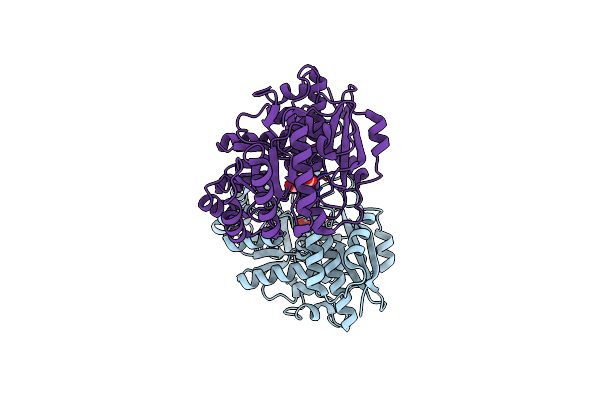 |
Crystal Structure Of Divergent Enolase From Oceanobacillus Iheyensis Complexed With Mg And L-Malate.
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-10-21 Classification: Isomerase, lyase Ligands: LMR, MG |
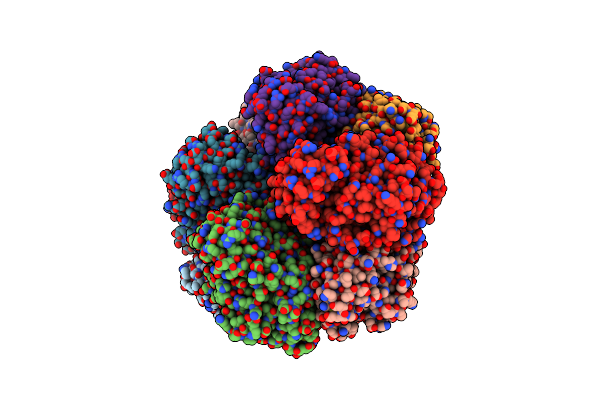 |
Crystal Structure Of Divergent Enolase From Oceanobacillus Iheyensis Complexed With Mg And L-Malate.
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-10-21 Classification: Isomerase, lyase Ligands: LMR, MG |
 |
Crystal Structure Of L-Rhamnonate Dehydratase From Salmonella Typhimurium Complexed With Mg And 3-Deoxy-L-Rhamnonate
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-09-23 Classification: LYASE Ligands: 3LR, MG, 1N5 |
 |
Crystal Structure Of Yfau, A Metal Ion Dependent Class Ii Aldolase From Escherichia Coli K12
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2008-09-09 Classification: LYASE Ligands: PO4, GOL |
 |
Crystal Structure Of Yfau, A Metal Ion Dependent Class Ii Aldolase From Escherichia Coli K12 - Mg-Pyruvate Product Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2008-09-09 Classification: LYASE Ligands: MG, PYR, PO4, GOL |
 |
Crystal Structure Of L-Rhamnonate Dehydratase From Salmonella Typhimurium Complexed With Mg
Organism: Salmonella typhimurium lt2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-01-01 Classification: LYASE Ligands: MG |
 |
Crystal Structure Of D-Mannonate Dehydratase From Novosphingobium Aromaticivorans
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-10-30 Classification: LYASE Ligands: MG |
 |
Crystal Structure Of The K271E Mutant Of Mannonate Dehydratase From Novosphingobium Aromaticivorans Complexed With Mg And D-Mannonate
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-10-30 Classification: LYASE Ligands: MG, CS2 |
 |
Crystal Structure Of D-Mannonate Dehydratase From Novosphingobium Aromaticivorans Complexed With Mg And 2-Keto-3-Deoxy-D-Gluconate
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-30 Classification: LYASE Ligands: MG, KDG |
 |
The Crystal Structure Of Muconate Cycloisomerase From Oceanobacillus Iheyensis
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-03-06 Classification: ISOMERASE Ligands: MG |
 |
Crystal Structure Of L-Fuconate Dehydratase From Xanthomonas Campestris Liganded With Mg++ And D-Erythronohydroxamate
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-12-19 Classification: UNKNOWN FUNCTION Ligands: MG, EHM |
 |
Crystal Structure Of K220A Mutant Of L-Fuconate Dehydratase From Xanthomonas Campestris Liganded With Mg++ And L-Fuconate
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-12-19 Classification: UNKNOWN FUNCTION Ligands: MG, SO4, LFC |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-09-05 Classification: LYASE |

