Search Count: 92
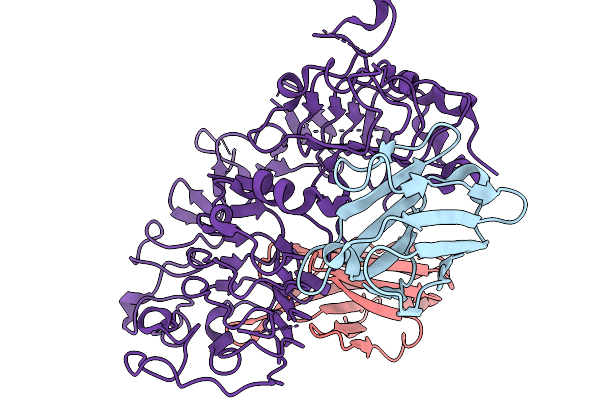 |
Cryoem Structure Of Extracellular Domain Of Human Her2 Complexed With Two Nano-Bodies 27A05 And 47D05
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSFERASE/IMMUNE SYSTEM |
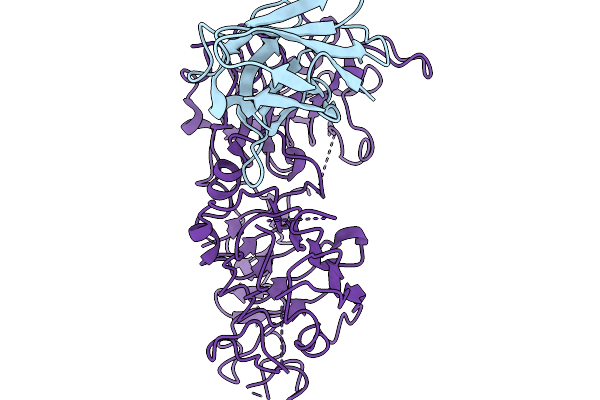 |
Cryoem Structure Of Extracellular Domain Of Human Her2 Complexed With Nano-Bodies 29E09
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSFERASE |
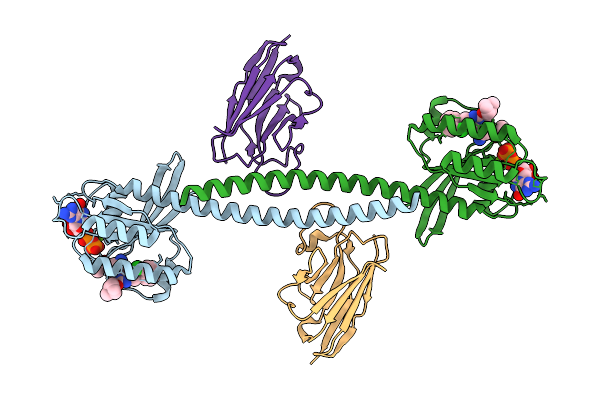 |
A Coiled Coil Module Strategy For High-Resolution Cryo-Em Structures Of Small Proteins For Drug Discovery
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN Ligands: M1X, GDP, MG |
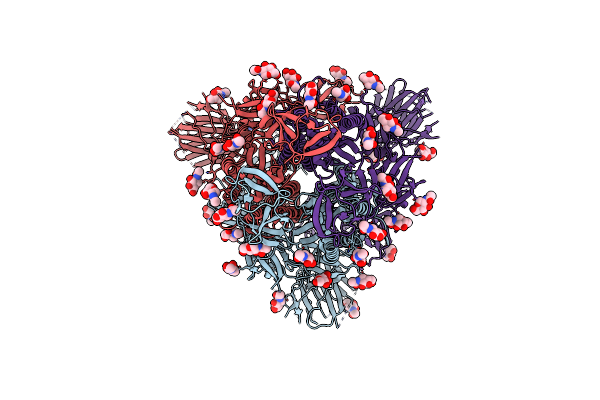 |
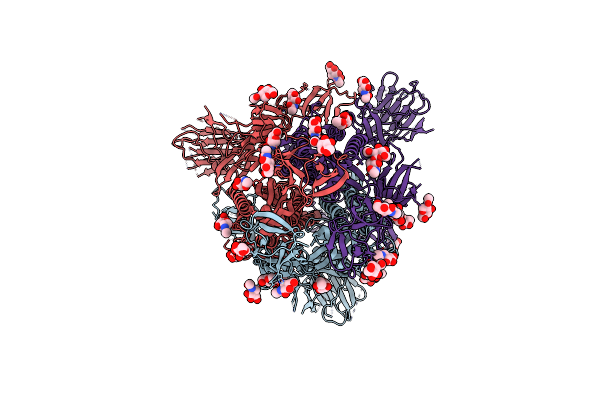
Structural And Biochemical Rationale For Beta Variant Protein Booster Vaccine Broad Cross-Neutralization Of Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
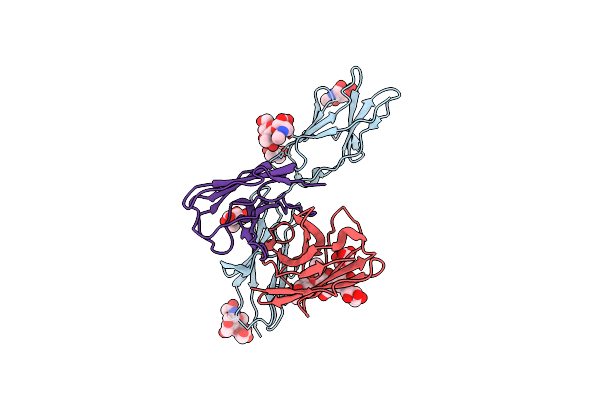
Structural And Biochemical Rationale For Beta Variant Protein Booster Vaccine Broad Cross-Neutralization Of Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Mus sp., Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: CELL ADHESION Ligands: NAG |
 |
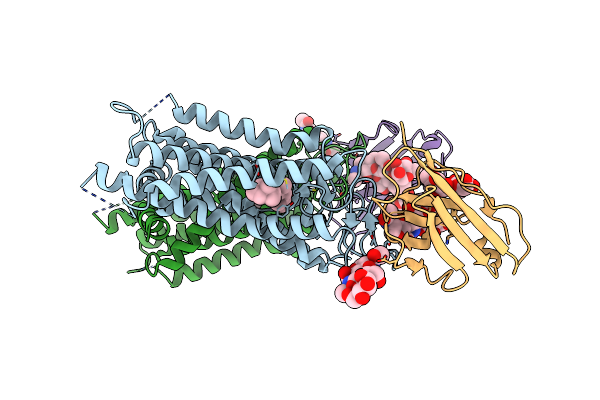
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2023-08-30 Classification: DNA BINDING PROTEIN Ligands: IHP |
 |
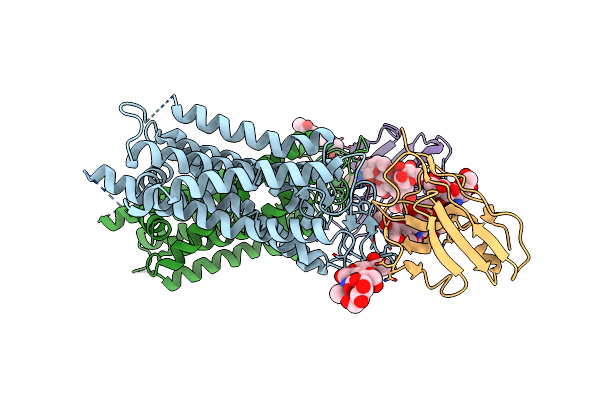
Organism: Lama glama, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Lama glama, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Lama glama, Bos taurus
Method: X-RAY DIFFRACTION Resolution:4.25 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: DNA BINDING PROTEIN Ligands: IHP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: DNA BINDING PROTEIN Ligands: IHP |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: RIBOSOME/ANTIBIOTIC Ligands: ZN, WDP, MG |
 |
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-06-08 Classification: RIBOSOME Ligands: MG, K, OHX, SJH |
 |
Organism: Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-06-01 Classification: RIBOSOME Ligands: MG, OHX, K, SJE |
 |
Organism: Sander vitreus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-05-25 Classification: FLUORESCENT PROTEIN Ligands: BLA, PG4 |
 |
Organism: Sander vitreus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-05-25 Classification: FLUORESCENT PROTEIN Ligands: BLA |
 |
Cryoem Structure Of A2296-Methylated Mycobacterium Tuberculosis Ribosome Bound With Seq-9
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RIBOSOME/ANTIBIOTIC Ligands: ZN, WDP, MG |
 |
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-06-03 Classification: TOXIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-11-27 Classification: TRANSCRIPTION Ligands: L5B |

