Search Count: 8
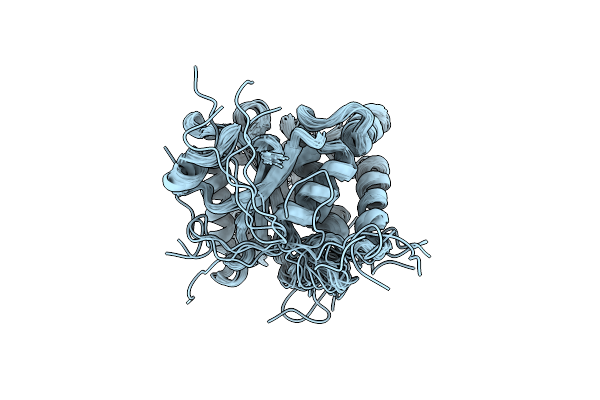 |
Multi-State Structure Determination And Dynamics Analysis Elucidate A New Ubiquitin-Recognition Mechanism Of Yeast Ubiquitin C-Terminal Hydrolase.
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2022-04-20 Classification: HYDROLASE |
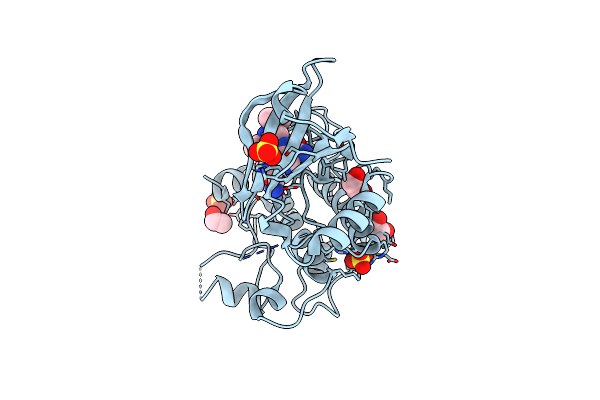 |
Crystal Structure Of Human Calmodulin-Dependent Protein Kinase 1D (Camk1D) Bound To Compound 19 (Cs640)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-11-13 Classification: TRANSFERASE Ligands: SO4, EDO, M92 |
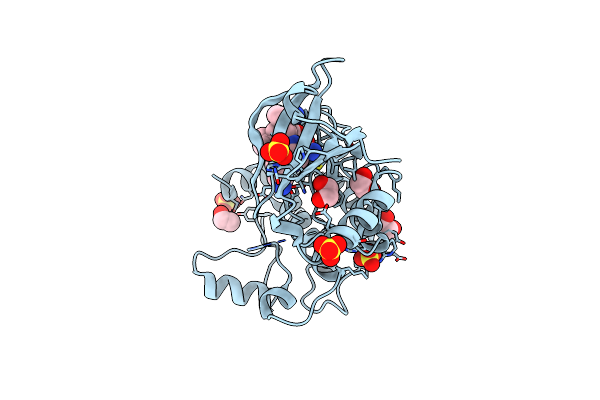 |
Crystal Structure Of Human Calmodulin-Dependent Protein Kinase 1D (Camk1D) Bound To Compound 18 (Cs587)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-11-13 Classification: TRANSFERASE Ligands: SO4, EDO, M8Z |
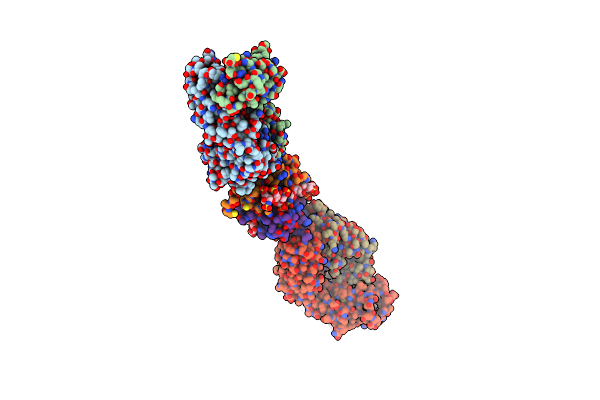 |
The Extracellular Domain Of G6B-B In Complex With Fab Fragment And Dp12 Heparin Oligosaccharide.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2019-09-04 Classification: BLOOD CLOTTING |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-05-04 Classification: PROTEIN TRANSPORT |
 |
Solution Structure Of Bame, A Component Of The Outer Membrane Protein Assembly Machinery In Escherichia Coli
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2011-02-02 Classification: MEMBRANE PROTEIN |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-03-13 Classification: OXIDOREDUCTASE Ligands: SO4, TXD, NAP |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2007-03-13 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, TXP |

