Search Count: 14
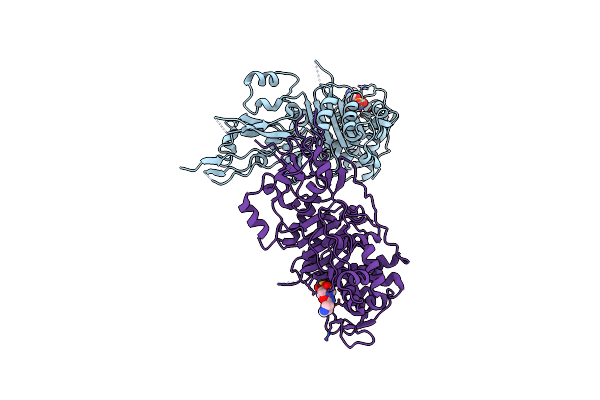 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2021-01-13 Classification: HYDROLASE/Inhibitor/Antibiotic Ligands: C9D, CL |
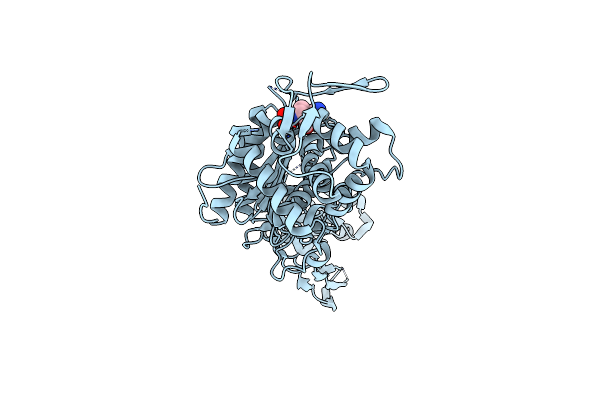 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-01-13 Classification: HYDROLASE/Inhibitor/Antibiotic Ligands: C8Y |
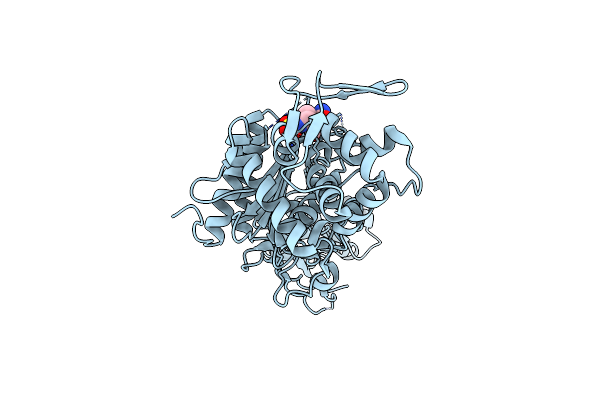 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2021-01-13 Classification: HYDROLASE/Inhibitor/Antibiotic Ligands: NXL |
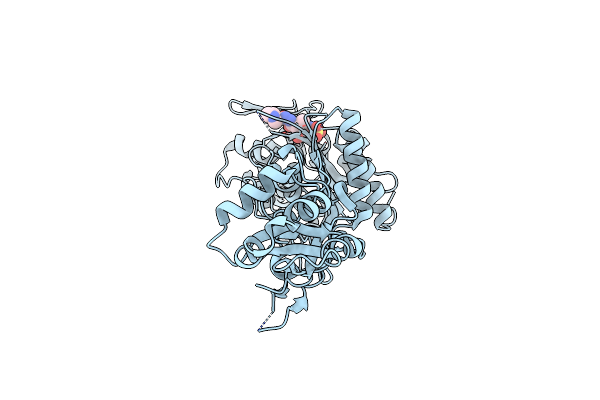 |
Crystal Structure Of Pseudomonas Aeruginosa Pbp3 In Complex With Zidebactam
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-01-13 Classification: HYDROLASE/Inhibitor/Antibiotic Ligands: C8V |
 |
Crystal Structure Of Lpqn Involved In Cell Envelope Biogenesis Of Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-09-18 Classification: LIPID BINDING PROTEIN |
 |
Crystal Structure Of Lpqn Involved In Cell Envelope Biogenesis Of Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-07-24 Classification: LIPID BINDING PROTEIN |
 |
Crystal Structure Of Lpqn Involved In Cell Envelope Biogenesis Of Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-07-24 Classification: LIPID BINDING PROTEIN Ligands: L6T |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Crystal Structure Of Ywfh, Nadph Dependent Reductase Involved In Bacilysin Biosynthesis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Ywfh, Nadph Dependent Reductase Involved In Bacilysin Biosynthesis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of Ywfh, Nadph Dependent Reductase Involved In Bacilysin Biosynthesis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE |
 |
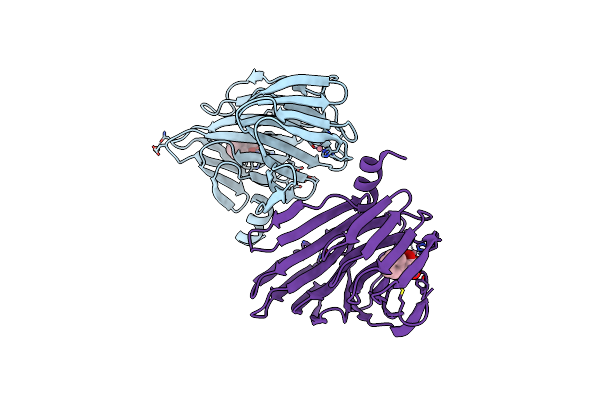
Crystal Structure Of Bacb, An Enzyme Involved In Bacilysin Synthesis, In Triclinic Form
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2010-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, PPY, FE |
 |
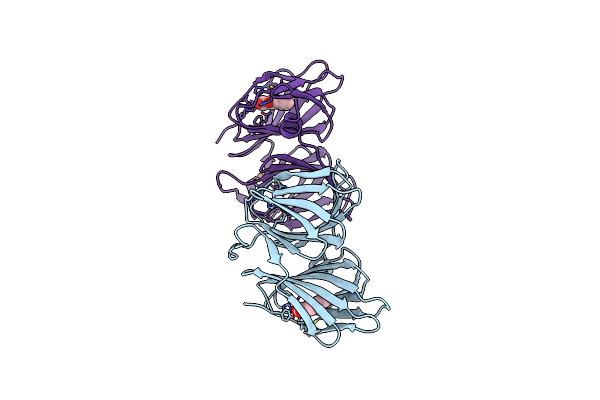
Crystal Structure Of Bacb, An Enzyme Involved In Bacilysin Synthesis, In Monoclinic Form
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2009-09-22 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, PPY, FE |
 |
Crystal Structure Of Bacb, An Enzyme Involved In Bacilysin Synthesis, In Tetragonal Form
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2009-09-22 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, PPY, FE |

