Search Count: 41
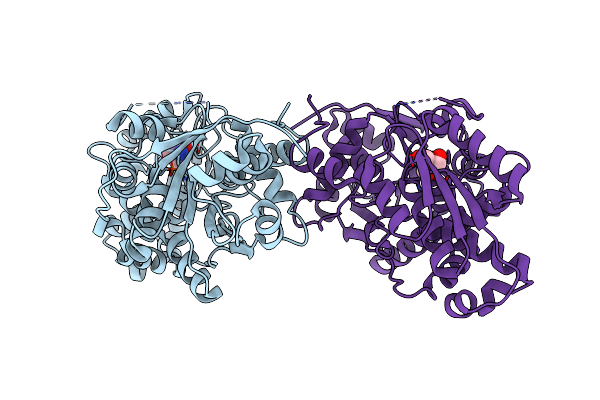 |
Crystal Structure Of Selenomethionine Incorporated Holo Diaminopropionate Ammonia Lyase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2012-04-25 Classification: LYASE Ligands: TRS |
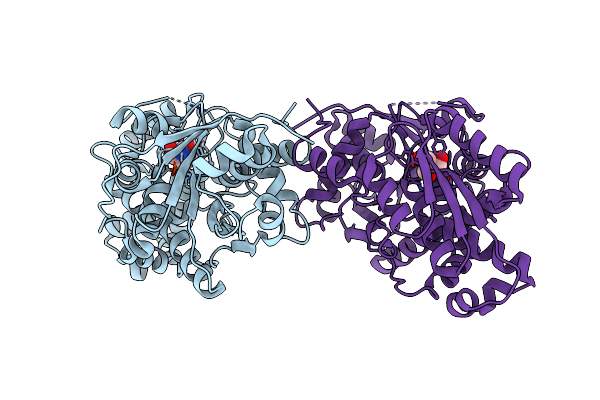 |
Crystal Structure Of Holo Diaminopropionate Ammonia Lyase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-04-25 Classification: LYASE Ligands: TRS |
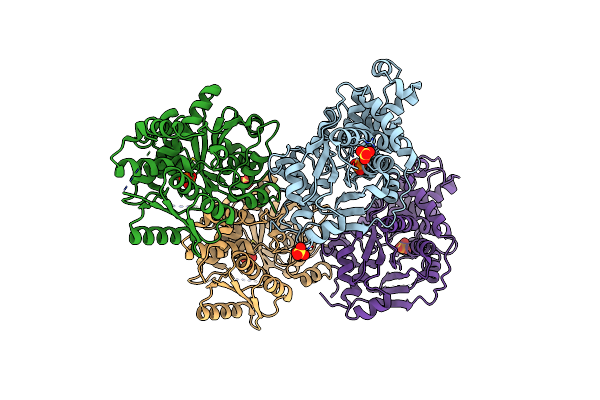 |
Crystal Structure Of Escherichia Coli Diaminopropionate Ammonia Lyase In Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2012-04-25 Classification: LYASE Ligands: PO4, SO4 |
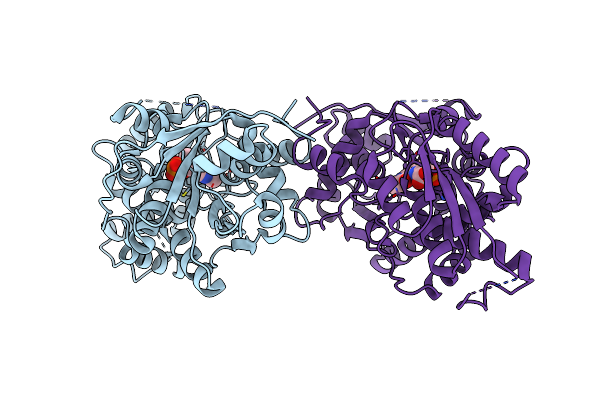 |
Crystal Structure Of Diaminopropionate Ammonia Lyase From Escherichia Coli In Complex With Aminoacrylate-Plp Azomethine Reaction Intermediate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-25 Classification: LYASE Ligands: 0JO |
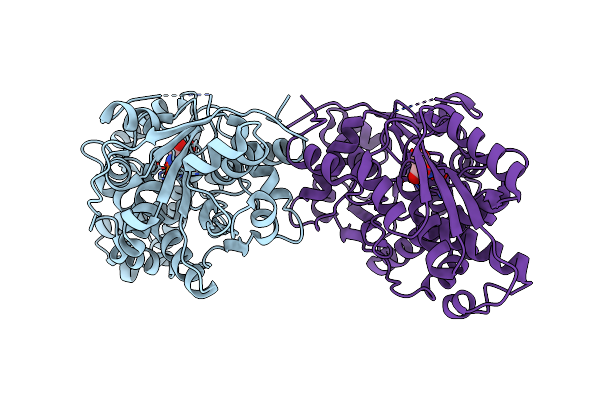 |
Crystal Structure Of Diaminopropionate Ammonia Lyase From Escherichia Coli In Complex With D-Serine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-25 Classification: LYASE Ligands: DSN |
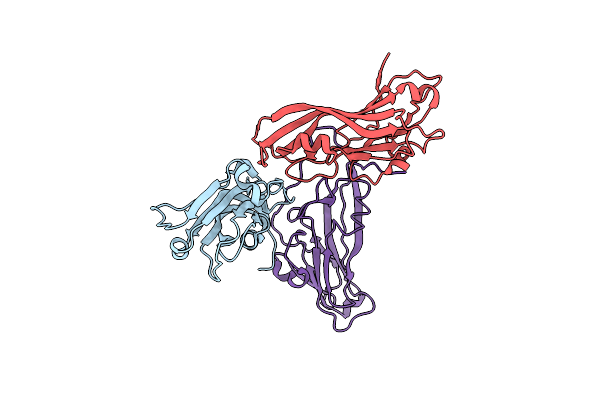 |
Organism: Physalis mottle virus
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2010-11-03 Classification: VIRUS |
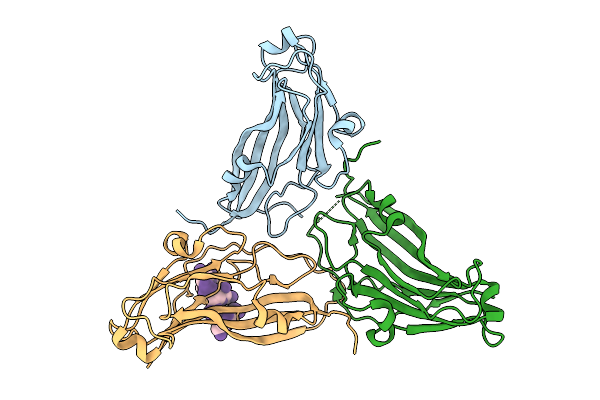 |
Organism: Physalis mottle virus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2010-09-15 Classification: VIRUS |
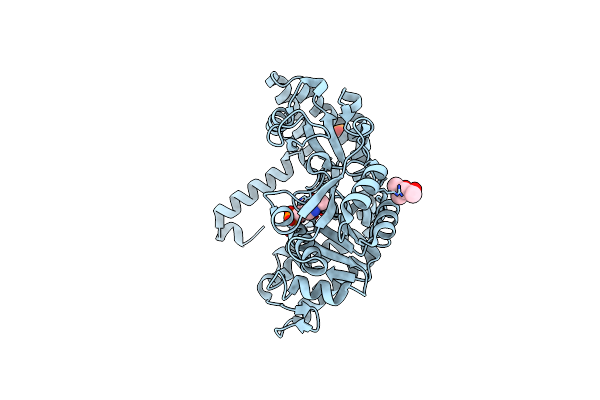 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: PLP, MPD, PO4 |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: GLY, PLP, MPD, PO4 |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: SER, PLP, MPD, PO4 |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: PLP, ALO, MPD, PO4 |
 |
Crystal Structure Of Y51Fbsshmt Obtained In The Presence Of Gly And 5- Formyl Tetrahydrofolate
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: GLY, PLP, MPD, PO4 |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: GLY, PLP |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: SER, PLP |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: PLP, ALO |
 |
Crystal Structure Of Y61Absshmt Obtained In The Presence Of Glycine And 5-Formyl Tetrahydrofolate
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2010-08-18 Classification: TRANSFERASE Ligands: GLY, PLP |
 |
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2008-12-16 Classification: TRANSFERASE Ligands: PLP, MPD, PO4 |
 |
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2008-12-16 Classification: TRANSFERASE Ligands: GLY, PLP, MPD, PO4 |
 |
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2008-12-16 Classification: TRANSFERASE Ligands: SER, PLP, MPD, PO4 |

