Search Count: 29
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-03-21 Classification: nuclear protein/antagonist Ligands: F3D, EDO, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-03-21 Classification: nuclear protein/antagonist Ligands: F3D, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-06-17 Classification: TRANSCRIPTION Ligands: JAD |
 |
Crystal Structure Of De Novo Designed Serine Hydrolase Osh18, Northeast Structural Genomics Consortium (Nesg) Target Or396
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-11-13 Classification: HYDROLASE |
 |
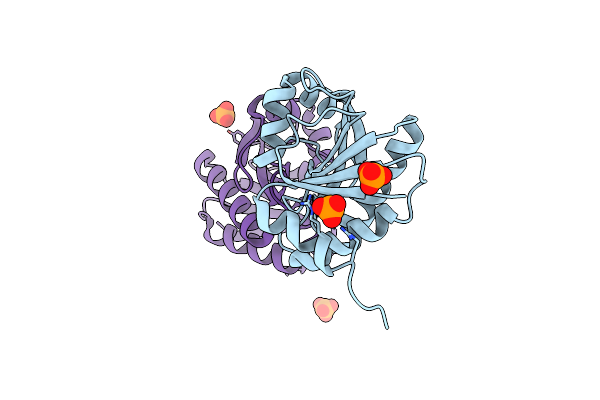
Crystal Structure Of Human Anaplastic Lymphoma Kinase In Complex With 7-Azaindole Based Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-07-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3DK |
 |
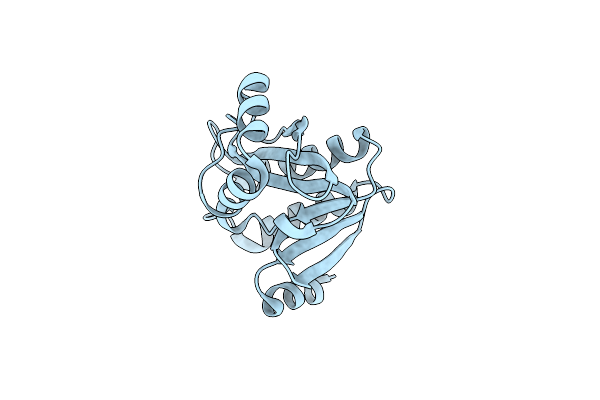
Crystal Structure Of De Novo Designed Serine Hydrolase Osh55.14_E3, Northeast Structural Genomics Consortium Target Or342
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2013-06-19 Classification: DE NOVO PROTEIN Ligands: PO4 |
 |
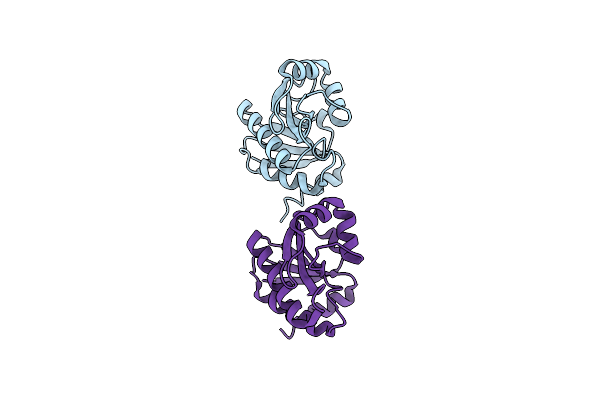
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-08 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2013-05-08 Classification: Transcription/DNA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2013-05-08 Classification: Transcription/DNA |
 |
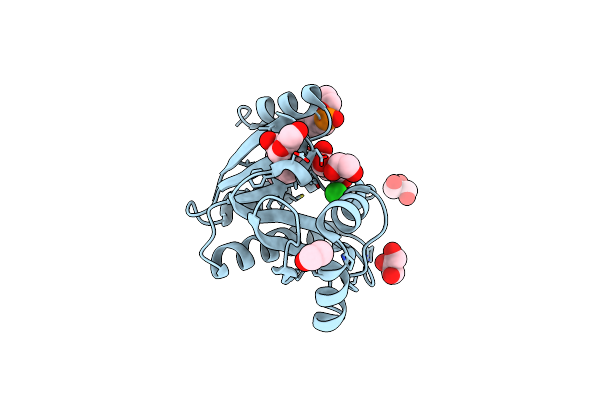
Crystal Structure Of The Evolved Variant Of The Computationally Designed Serine Hydrolase, Osh55.4_H1, Covalently Bound With Diisopropyl Fluorophosphate (Dfp), Northeast Structural Genomics Consortium (Nesg) Target Or273
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2013-04-24 Classification: Structural Genomics, Unknown Function |
 |
Crystal Structure Of The Computationally Designed Serine Hydrolase. Northeast Structural Genomics Consortium (Nesg) Target Or317
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-04-24 Classification: Structural Genomics, Unknown Function |
 |
Crystal Structure Of The Evolved Variant Of The Computationally Designed Serine Hydrolase, Osh55.4_H1 Covalently Bound With Fp-Alkyne, Northeast Structural Genomics Consortium (Nesg) Target Or273
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2013-04-10 Classification: Structural Genomics, Unknown Function Ligands: SEF, CL, EDO, PEG, GOL |
 |
Crystal Structure Of The Computationally Designed Serine Hydrolase 3Mmj_2, Northeast Structural Genomics Consortium (Nesg) Target Or318
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-03-20 Classification: Structural Genomics, Unknown Function Ligands: PO4 |
 |
Crystal Structure Of The Apo Form Of The Evolved Variant Of The Computationally Designed Serine Hydrolase, Osh55.4_H1. Northeast Structural Genomics Consortium (Nesg) Target Or273
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2013-03-20 Classification: HYDROLASE Ligands: RB, CIT |
 |
Crystal Structure Of The Evolved Variant Of The Computationally Designed Serine Hydrolase, Northeast Structural Genomics Consortium (Nesg) Target Or275
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-03-20 Classification: Structural Genomics, Unknown Function |
 |
Crystal Structure Of The Improved Variant Of The Evolved Serine Hydrolase, Osh55.4_H1.2, Bond With Sulfate Ion In The Active Site, Northeast Structural Genomics Consortium (Nesg) Target Or301
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2013-03-06 Classification: Structural Genomics, Unknown Function Ligands: SO4, PEG, EPE |
 |
Crystal Structure Of De Novo Design Serine Hydrolase Osh55.27, Northeast Structural Genomics Consortium (Nesg) Target Or246
Method: X-RAY DIFFRACTION
Resolution:2.90 Å Release Date: 2012-09-12 Classification: DE NOVO PROTEIN |
 |
Three-Dimensional Structure Of The De Novo Designed Serine Hydrolase 2Bfq_3, Northeast Structural Genomics Consortium (Nesg) Target Or248
Method: X-RAY DIFFRACTION
Resolution:2.11 Å Release Date: 2012-09-12 Classification: DE NOVO PROTEIN, HYDROLASE Ligands: PO4, MES, ACY |
 |
Crystal Structure Of E6D/L155R Variant Of De Novo Designed Serine Hydrolase Osh55, Northeast Structural Genomics Consortium (Nesg) Target Or187
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: De Novo Protein, hydrolase |
 |
Crystal Structure Of E6H Variant Of De Novo Designed Serine Hydrolase Osh55, Northeast Structural Genomics Consortium (Nesg) Target Or185
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-06-13 Classification: DE NOVO PROTEIN Ligands: CL, PE4, PE5, PE6 |

