Search Count: 37
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: SIGNALING PROTEIN |
 |
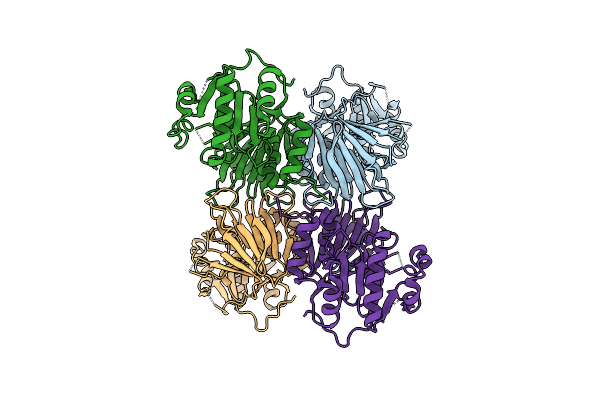
Cryoem Structure Of The C-Terminally Truncated Form Of Human Nad Kinase Bound To Nad
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: SIGNALING PROTEIN Ligands: NAD |
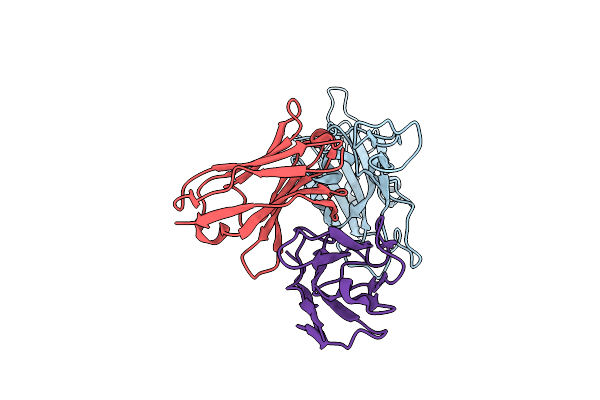 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
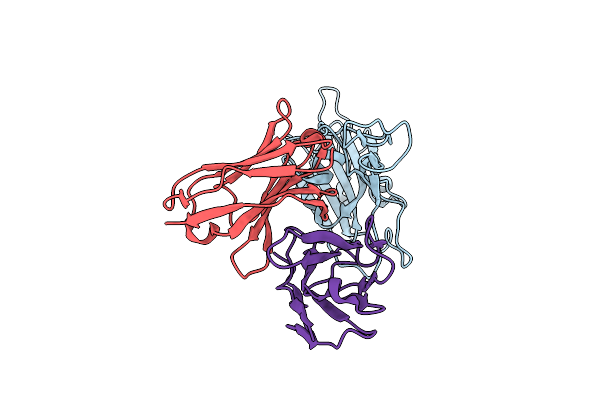 |
Model And Map From Local Refinement Of A Cab-A17 - Omicron Ba.1 Spike Complex
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
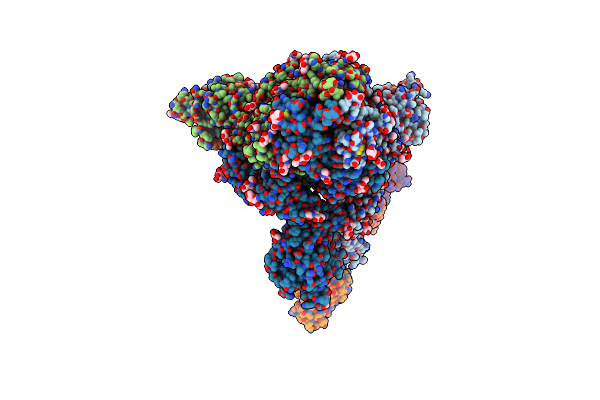 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2023-07-12 Classification: HYDROLASE Ligands: BR8, CIT, EDO, NA |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO, CL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Barbituric Acid, Form Ii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: BR8, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Isoorotic Acid (2,4-Dihydroxypyrimidine-5-Carboxylic Acid), Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: 5CU, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Isoorotic Acid (2,4-Dihydroxypyrimidine-5-Carboxylic Acid) And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: 5CU, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 2,4-Thiazolidinedione, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: OD3, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Uric Acid, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: URC, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 5-Fluoroorotic Acid And Citric Acid, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: FOT, CIT, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With Orotic Acid, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: ORO, EDO |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 5-Hydroxy-2,4(1H,3H)-Pyrimidinedione, Form I
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: ODF, EDO, DMS, NA, CL |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 6-Formyl-Uracil, Form Iii
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: OG6 |
 |
Crystal Structure Of Mycobacterium Tuberculosis Uracil-Dna Glycosylase In Complex With 5-Hydroxy-2,4(1H,3H)-Pyrimidinedione, Form Vi
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-07-12 Classification: HYDROLASE/INHIBITOR Ligands: ODF, EDO, PEG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN Ligands: NAG |

