Search Count: 15
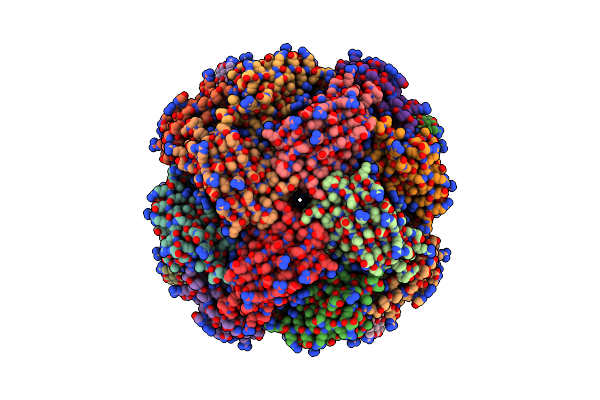 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
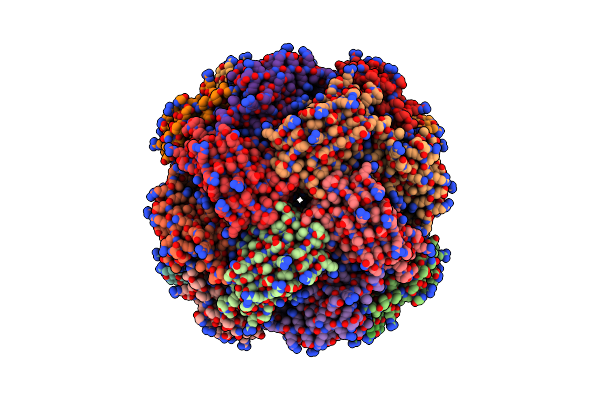 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, In Complex With Aminotriazole
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN, 3TR |
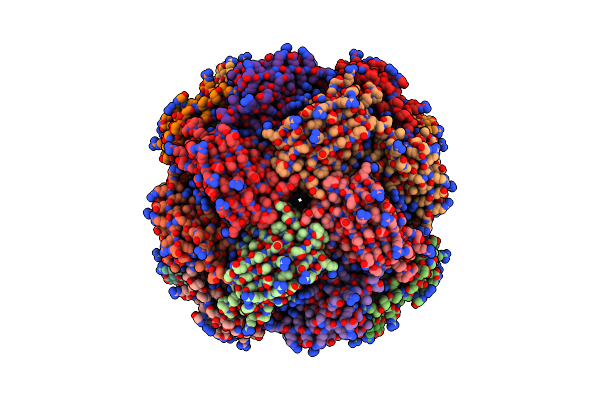 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168), Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: RIBOSOME Ligands: ZN |
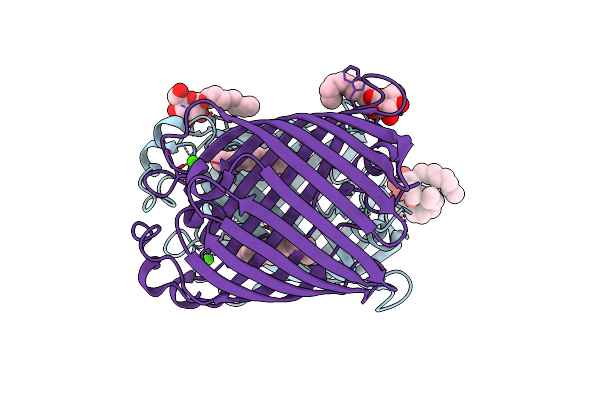 |
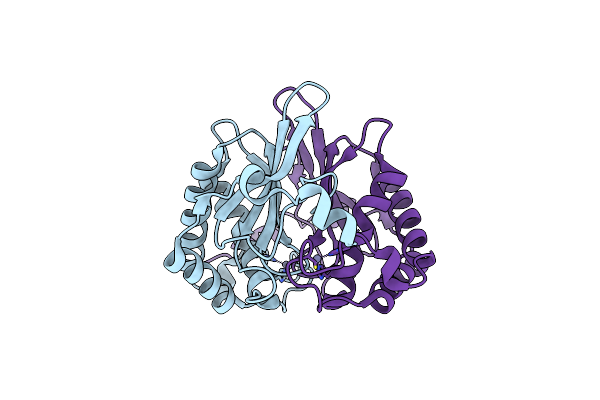
Crystal Structure Of Salmonella Typhi Outer Membrane Phospholipase (Ompla) Dimer With Bound Calcium
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: BOG, CA, D12 |
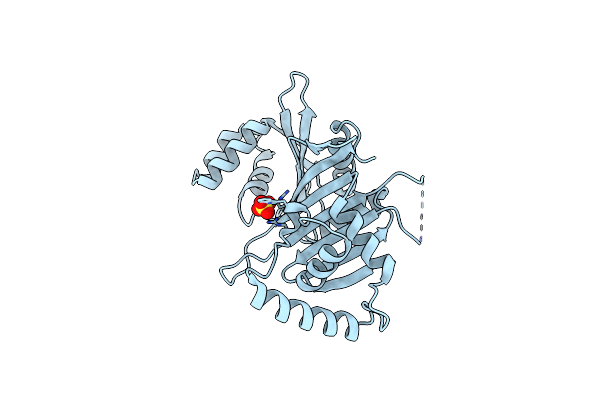 |
Crystal Structure Of Rv0289/Espg3 From The Esx-3 Type Vii Secretion System Of M. Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2018-05-16 Classification: CHAPERONE Ligands: SO4 |
 |
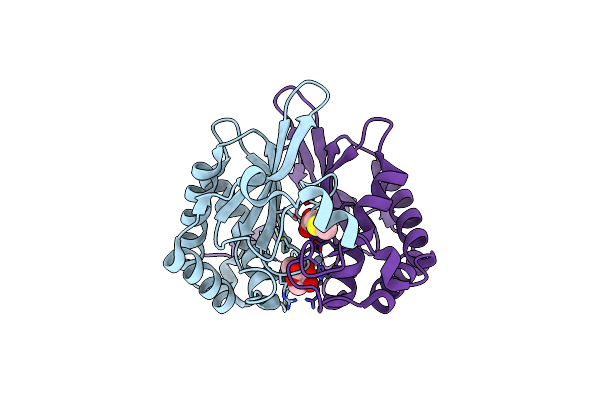
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-23 Classification: LYASE Ligands: ZN |
 |
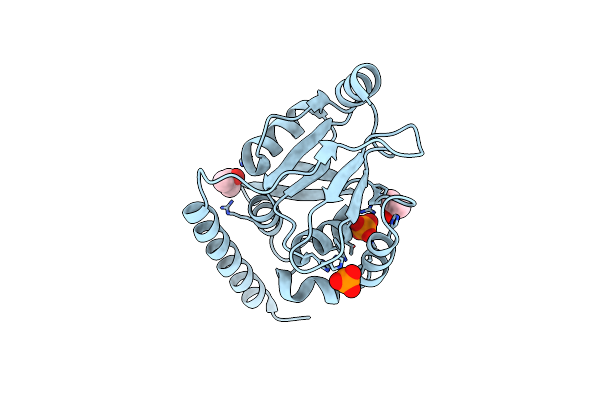
High Resolution Crystal Structure Of Luxs - Quorum Sensor Molecular Complex From Salmonella Typhi At 1.58 Angstroms
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2016-10-12 Classification: LYASE Ligands: ZN, PAV, MET |
 |
Crystal Structure Of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-11-05 Classification: ANTITUMOR PROTEIN Ligands: PO4, CL, IPA |
 |
Crystal Structure Of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor, Gold (I) Cyanide Derivative
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2014-11-05 Classification: ANTITUMOR PROTEIN Ligands: PO4, AUC, GOL |

