Search Count: 15
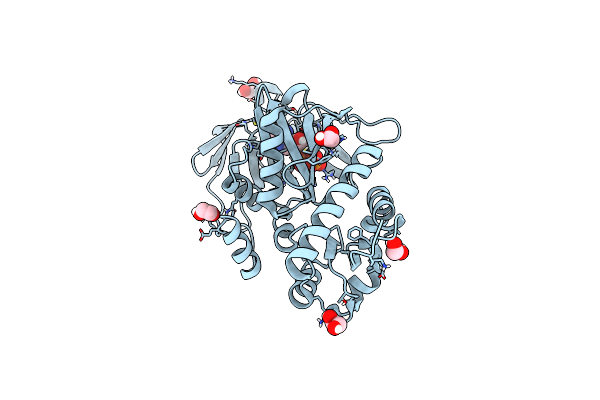 |
Crystal Structure Of The N-Terminal Kinase Domain From Saccharomyces Cerevisiae Vip1 In Complex With Adp.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, EDO |
 |
Crystal Structure Of The C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 (Apo, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, ZN, LPC, ACT |
 |
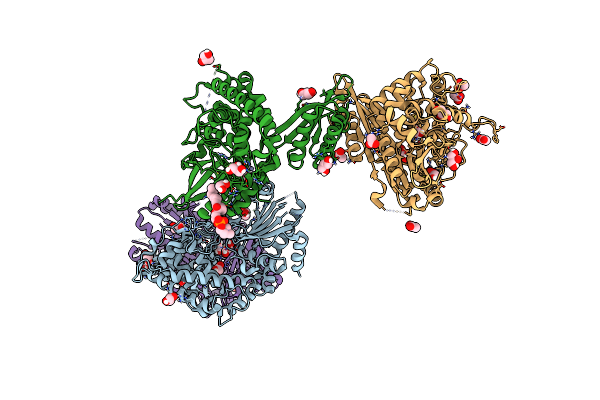
Crystal Structure Of The Engineered C-Terminal Phosphatase Domain From Saccharomyces Cerevisiae Vip1 In Complex With 1,5-Insp8 (Phosphatase Dead Mutant, Loop Deletion Residues 848-918)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-02-26 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, SPM, ORN, PUT, EDO, I8P, 1JW |
 |
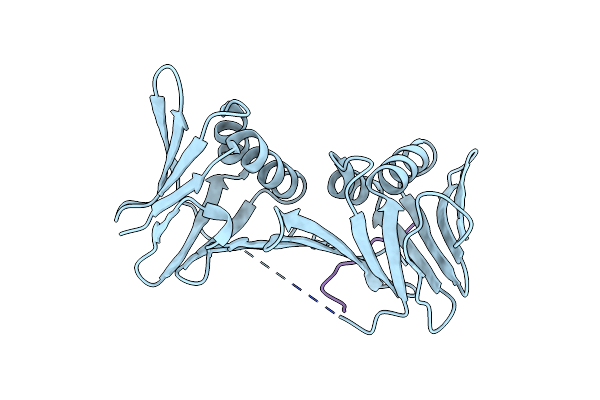
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-09-11 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: HYDROLASE |
 |
Organism: Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-03-04 Classification: REPLICATION |
 |
Organism: Pyrococcus abyssi (strain ge5 / orsay), Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-03-04 Classification: REPLICATION |
 |
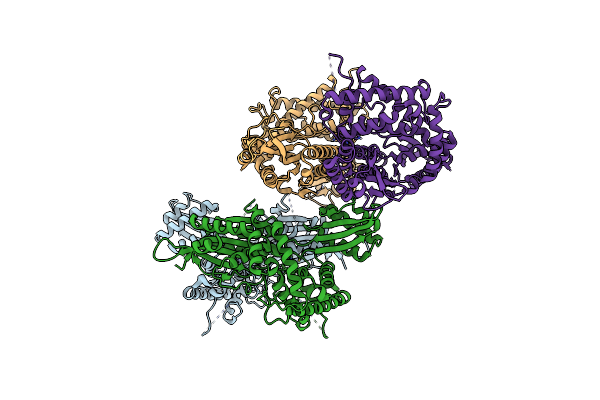
Cryo-Em Structure Of The Dna-Bound Pold-Pcna Processive Complex From P. Abyssi
Organism: Pyrococcus abyssi (strain ge5 / orsay), Pyrococcus abyssi, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-03-04 Classification: REPLICATION Ligands: FE, ZN |
 |
D-Family Dna Polymerase - Dp1 Subunit (3'-5' Proof-Reading Exonuclease) H451 Proof-Reading Deficient Variant
Organism: Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-01-30 Classification: REPLICATION Ligands: FE, ZN, ACT, CAC, CA |
 |
Organism: Pyrococcus abyssi
Method: ELECTRON MICROSCOPY Release Date: 2019-01-30 Classification: REPLICATION Ligands: FE, ZN |
 |
Organism: Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-31 Classification: TRANSFERASE Ligands: D5M, FE, ZN, CA, ACT, EDO |
 |
Organism: Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2016-08-31 Classification: TRANSFERASE Ligands: ZN |

