Search Count: 96
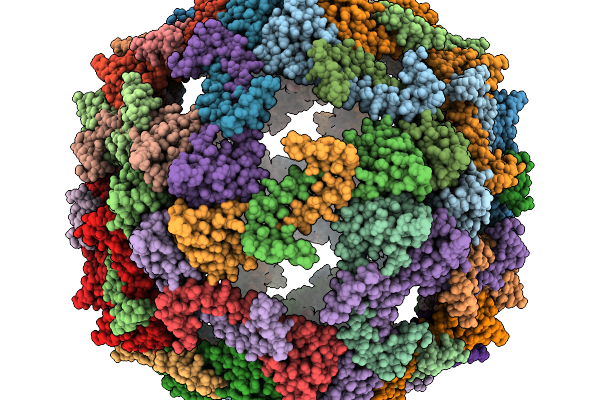 |
Cryo-Em Structure Of Pyrene-Modified Tip60 Double Mutant (G12C/S50C) With Addition Of Nile Red
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: A1L9F |
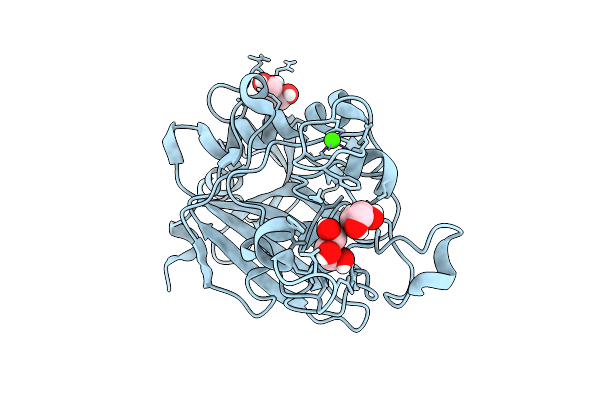 |
Crystal Structure Of Wild-Type Human Fibrinogen Gamma Chain C-Terminal Domain (Gamma-Nodule)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BLOOD CLOTTING Ligands: GOL, CA |
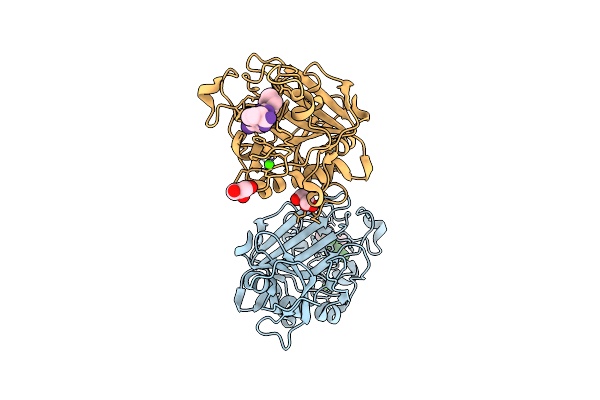 |
Crystal Structure Of Wild-Type Human Fibrinogen Gamma Chain C-Terminal Domain (Gamma-Nodule) Complexed With Gprp Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BLOOD CLOTTING Ligands: CA, GOL |
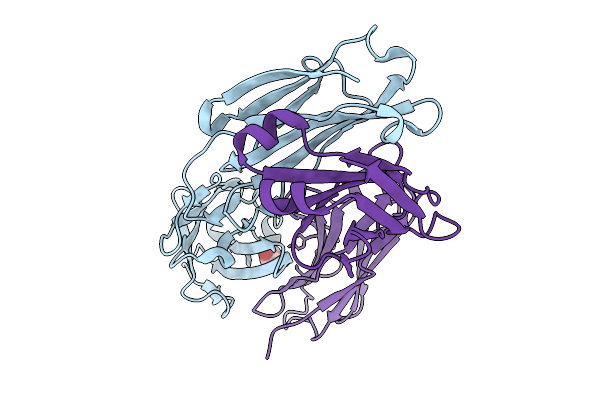 |
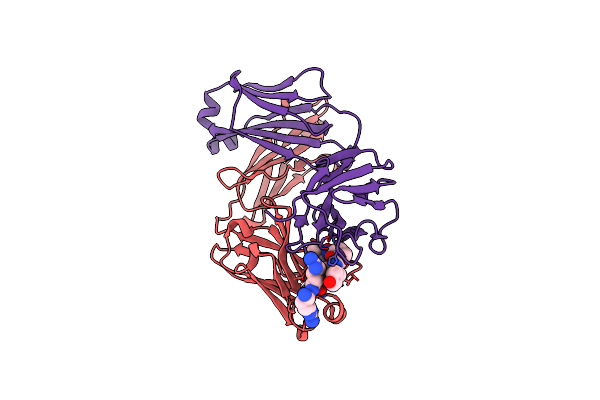
Crystal Structure Of Switchbody Based On Anti-Osteocalcin Antibody Ktm219 Fab
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
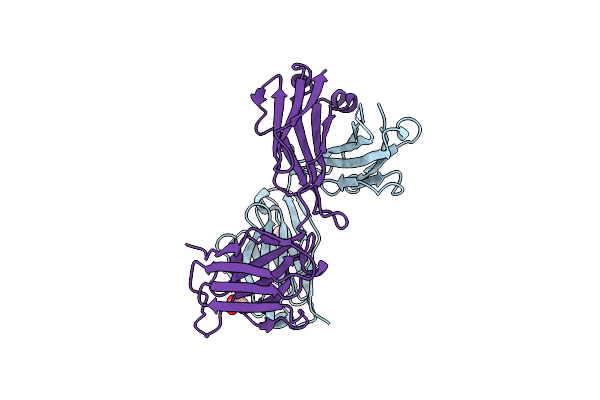
Crystal Structure Of Fab Fragment Of Anti-Osteocalcin Antibody Ktm219 Complexed With C-Terminal Peptide Antigen
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-15 Classification: IMMUNE SYSTEM |
 |
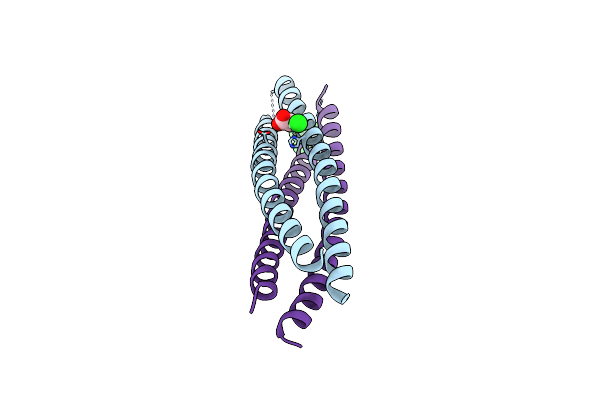
Crystal Structure Of Fab Fragment Of Anti-Osteocalcin C-Terminal Peptide Antibody Ktm219 With Addition Of Fluorophore Tamra
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-01-15 Classification: IMMUNE SYSTEM Ligands: ACT |
 |
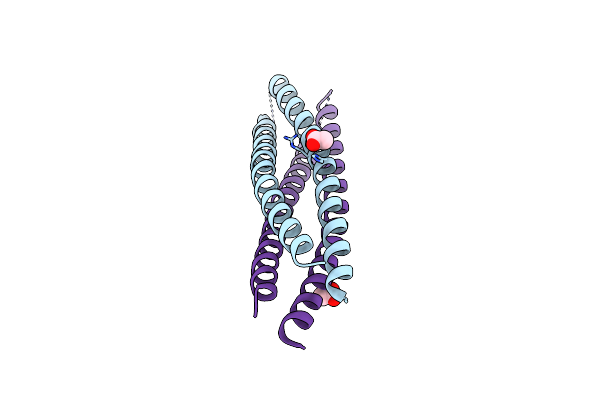
Crystal Structure Of A De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 (K4T) - Pt Derivative
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-09-20 Classification: DE NOVO PROTEIN Ligands: PT, CL, ACT, PC4 |
 |
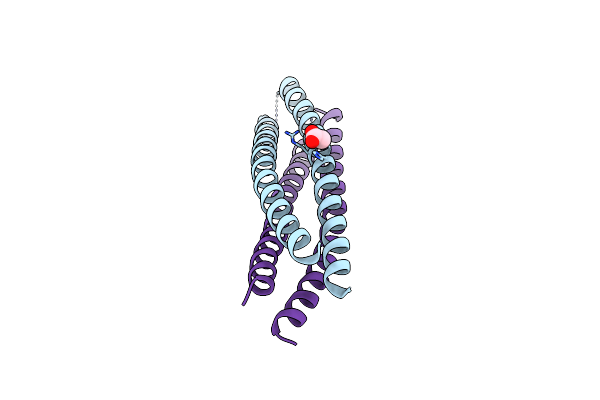
Crystal Structure Of A De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 (K4T)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-20 Classification: DE NOVO PROTEIN Ligands: ACT |
 |
Crystal Structure Of A De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 (K4T) At 2.0 Angstrom Resolution
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-09-20 Classification: DE NOVO PROTEIN Ligands: ACT |
 |
Cryo-Em Structure Of Mtip60-Ba (Metal-Ion Induced Tip60 (K67E) Complex With Barium Ions
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: DE NOVO PROTEIN Ligands: BA |
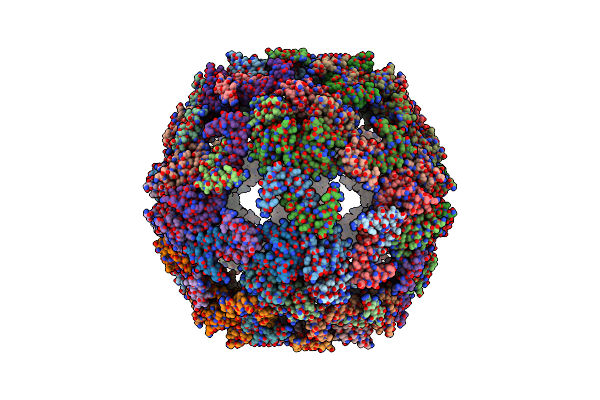 |
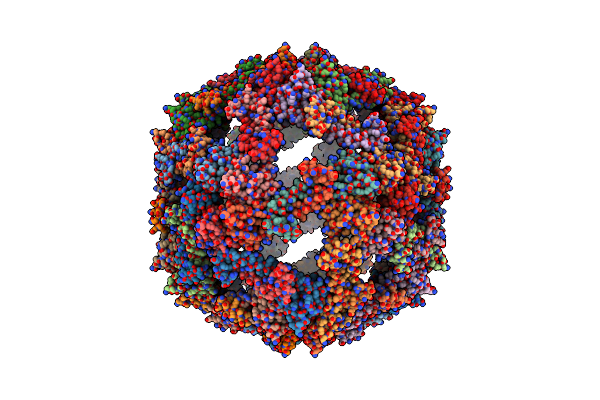
Cryo-Em Structure Of Designed Protein Nanoparticle Tip60 (Truncated Icosahedral Protein Composed Of 60-Mer Fusion Proteins)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-09-15 Classification: DE NOVO PROTEIN |
 |
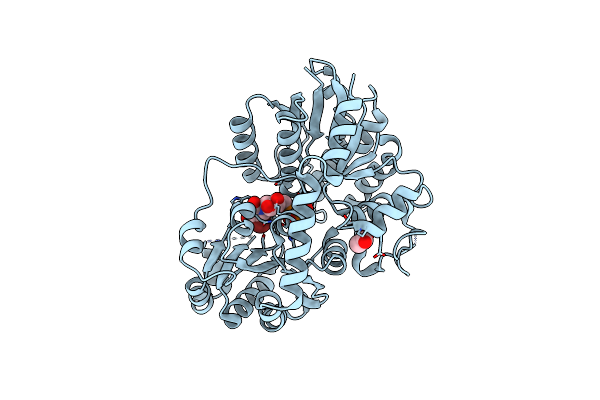
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-10-21 Classification: PROTEIN BINDING Ligands: GOL |
 |
Crystal Structure Of A Flavonoid C-Glucosyltrasferase From Fagopyrum Esculentum (Fecgta) Complexed With Brutp
Organism: Fagopyrum esculentum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: ACT, BUP |
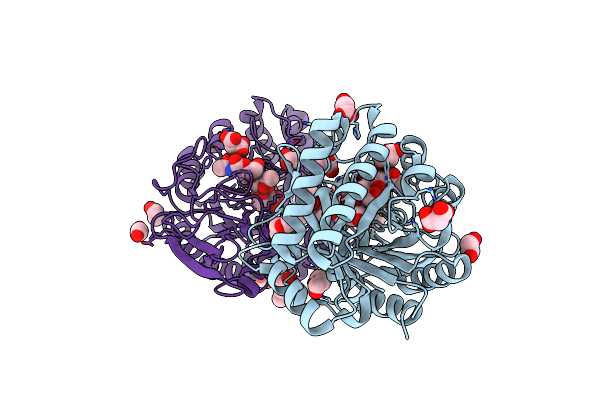 |
Crystal Structure Of The Catalytic Domain Of Chitiniphilus Shinanonensis Chitinase Chil (Cschil) Complexed With N,N'-Diacetylchitobiose
Organism: Chitiniphilus shinanonensis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: ACT, MXE, MLI, PGE, EDO, NAG, PG4 |
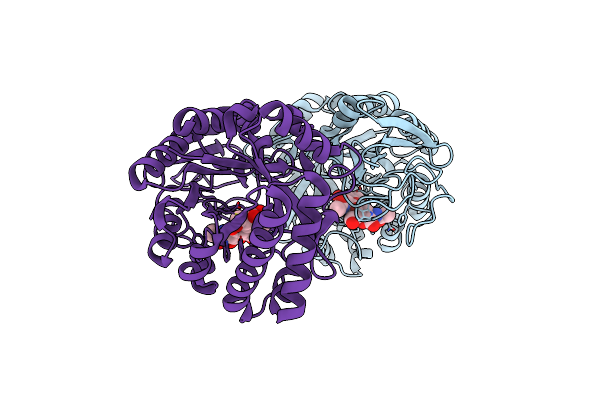 |
Crystal Structure Of D157N Mutant Of Chitiniphilus Shinanonensis Chitinase Chil (Cschil) Complexed With N,N'-Diacetylchitobiose
Organism: Chitiniphilus shinanonensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-09-16 Classification: HYDROLASE |
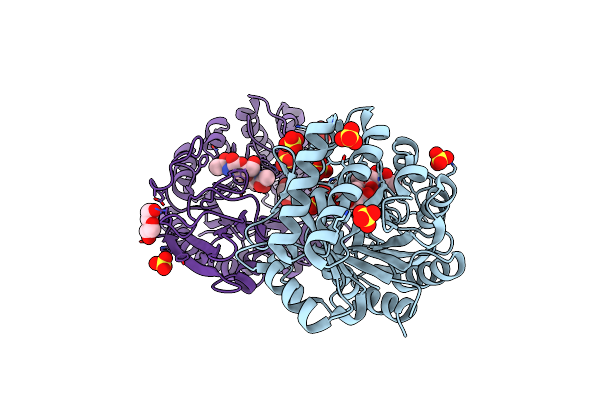 |
Crystal Structure Of W50A Mutant Of Chitiniphilus Shinanonensis Chitinase Chil (Cschil) Complexed With N,N'-Diacetylchitobiose
Organism: Chitiniphilus shinanonensis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: SO4, PEG, GOL, EDO |
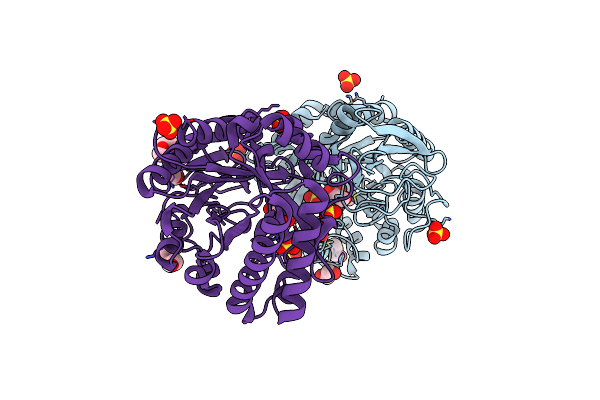 |
Crystal Structure Of The Catalytic Domain Of Chitinase Chil From Chitiniphilus Shinanonensis (Cschil)
Organism: Chitiniphilus shinanonensis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-08-26 Classification: HYDROLASE Ligands: GOL, SO4, EDO, MXE, CL |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-06 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-05-06 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN |
 |
Crystal Structure Of Suwa (Super Wa20), A Hyper-Stable De Novo Protein With A Dimeric Bisecting Topology
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-29 Classification: DE NOVO PROTEIN |

