Search Count: 19
 |
Crystal Structure Of Hyperthermostable Carboxylesterase From Anoxybacillus Geothermalis D9
Organism: Anoxybacteroides rupiense
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Organism: Photobacterium sp. j15
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-01-17 Classification: HYDROLASE |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: ZN, CA, NA, CL |
 |
Unravelling Structure Of Riboflavin Synthase For Designing Of Potential Anti-Bacterial Drug
Organism: Leptospira kmetyi serovar malaysia str. bejo-iso9
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2022-06-15 Classification: ANTIMICROBIAL PROTEIN Ligands: PEG |
 |
Organism: Bacillus lehensis g1
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: ZN, IOD |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2021-04-07 Classification: HYDROLASE Ligands: ZN, CA |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-23 Classification: METAL BINDING PROTEIN |
 |
Organism: Photobacterium sp. j15(2011)
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: PEG, CL, CAC, CA, EDO, PO4 |
 |
Structure Elucidation Of Truncated Ams3 Lipase From An Antarctic Pseudomonas
Organism: Pseudomonas sp. a3(2015c)
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Pseudomonas Aeruginosa Strain K Solvent Tolerant Elastase
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: GOL, PO4, CA, ZN |
 |
Crystal Structure Of Thermostable, Organic-Solvent Tolerant Lipase From Geobacillus Sp. Strain Arm
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-07-31 Classification: HYDROLASE Ligands: ZN, CA |
 |
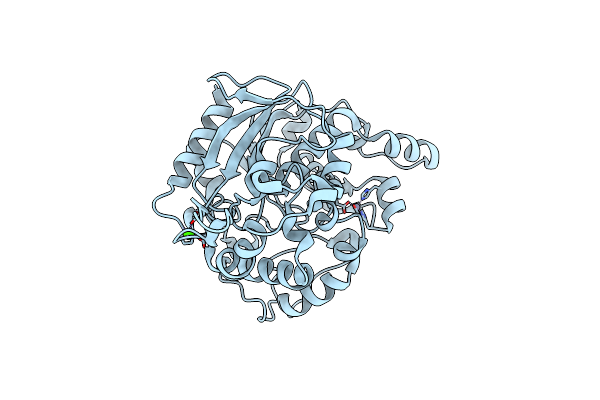
Organism: Bacillus sp. 42
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2013-06-19 Classification: HYDROLASE Ligands: ZN, GOL, NA, CL, CA |
 |
Crystallization And 3D Structure Elucidation Of Thermostable L2 Lipase From Thermophilic Locally Isolated Bacillus Sp. L2.
Organism: Bacillus sp. l2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: ZN, CA |
 |
 |
 |
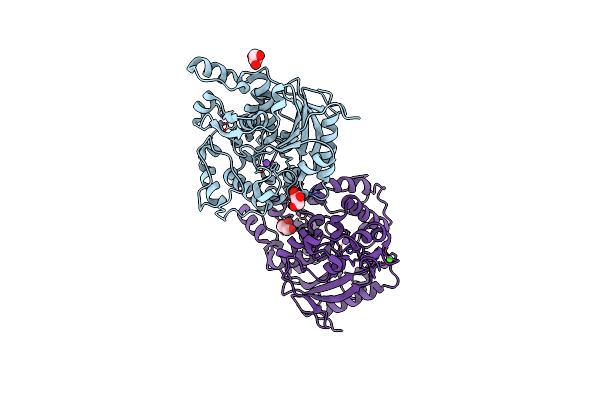 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: ZN, GOL, CL, NA, CA |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-10-30 Classification: HYDROLASE Ligands: ZN, CA, CL |
 |
Organism: Geobacillus zalihae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-07-17 Classification: HYDROLASE Ligands: ZN, CA, NA, CL |

