Search Count: 95
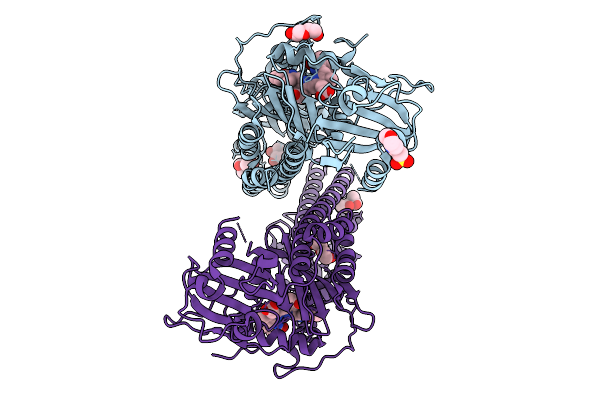 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf205 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, MPD, PEG, MES, CL |
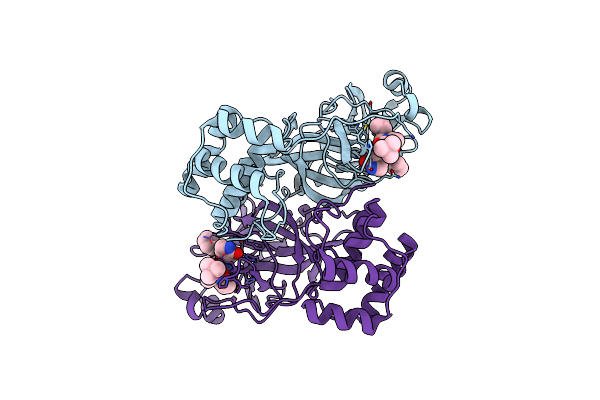 |
Sars-Cov-2 Mpro (Omicron, P132H+T169S) In Complex With Alpha-Ketoamide 13B-K
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-11-13 Classification: VIRAL PROTEIN Ligands: O6K |
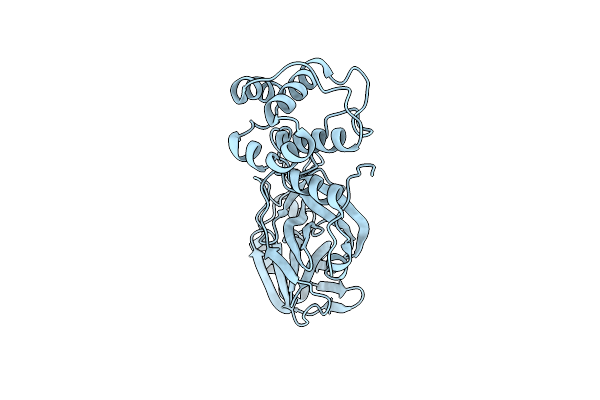 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-11-13 Classification: VIRAL PROTEIN |
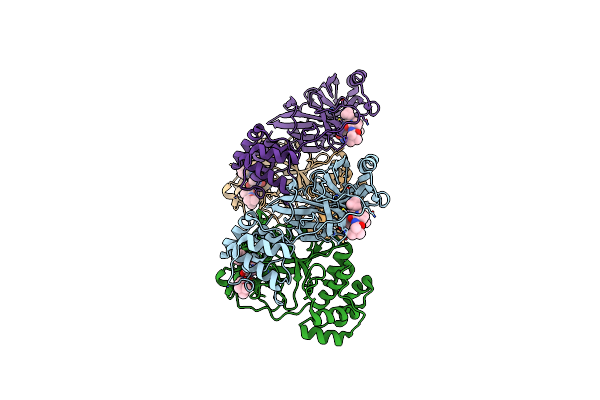 |
Sars-Cov-2 Mpro (Omicron,P132H) In Complex With Alpha-Ketoamide 13B-K At Ph 8.5
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-13 Classification: VIRAL PROTEIN Ligands: O6K, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-07 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-07 Classification: ANTIVIRAL PROTEIN Ligands: CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-08-07 Classification: ANTIVIRAL PROTEIN Ligands: O6K, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-02-07 Classification: ANTIVIRAL PROTEIN |
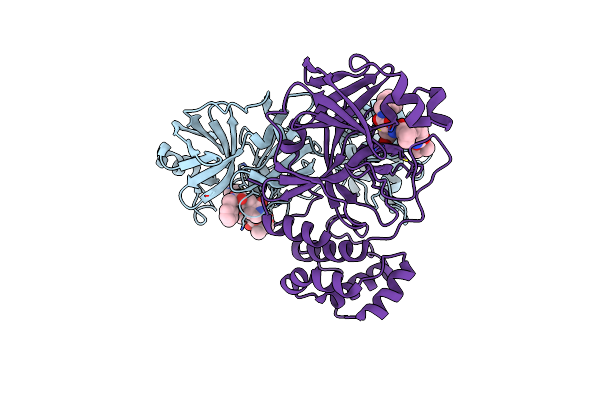 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-02-07 Classification: ANTIVIRAL PROTEIN Ligands: O6K |
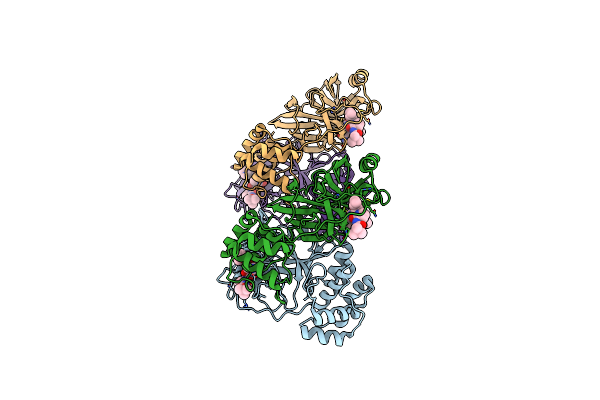 |
Sars-Cov-2 Mpro (Omicron, P132H) In Complex With Alpha-Ketoamide 13B-K At Ph 6.5
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN Ligands: O6K |
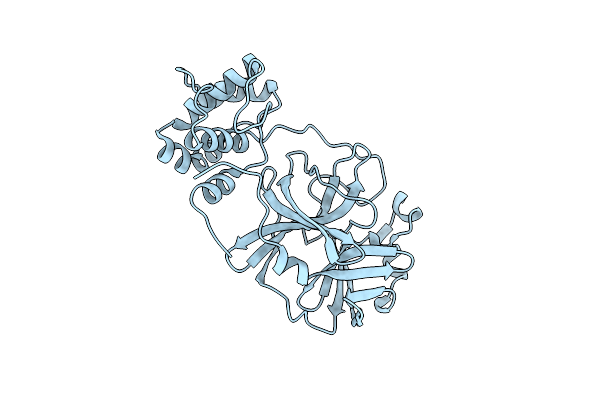 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-11-29 Classification: VIRAL PROTEIN |
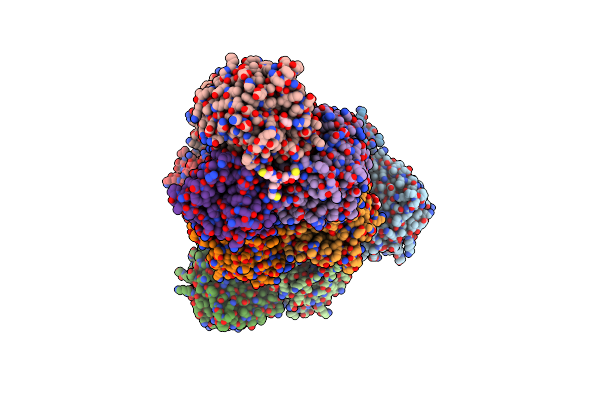 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZIZ, GOL |
 |
Monomer Structure Of Transforming Growth Factor Beta Induced Protein (Tgfbip) G623R Fibril
Organism: Homo sapiens
Method: SOLID-STATE NMR Release Date: 2023-07-26 Classification: PROTEIN FIBRIL |
 |
Structure Of Transforming Growth Factor Beta Induced Protein (Tgfbip) G623R Fibril
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: PROTEIN FIBRIL |
 |
Rt Xfel Structure Of The Two-Flash State Of Photosystem Ii (2F, S3-Rich) At 2.09 Angstrom Resolution
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-03-22 Classification: PHOTOSYNTHESIS Ligands: OEY, OEX, FE2, CL, CLA, PHO, BCR, PL9, LMG, SQD, LHG, DGD, STE, BCT, HEM, HEC |
 |
Rt Xfel Structure Of The Two-Flash State Of Photosystem Ii (2F, S3-Rich) At 2.00 Angstrom Resolution
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-22 Classification: PHOTOSYNTHESIS Ligands: OEY, OEX, FE2, CL, CLA, BCR, PL9, LMG, LHG, SQD, DGD, STE, BCT, PHO, HEM, HEC |
 |
Rt Xfel Structure Of Photosystem Ii 50 Microseconds After The Third Illumination At 2.15 Angstrom Resolution
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-03-22 Classification: PHOTOSYNTHESIS Ligands: OEY, OEX, FE2, CL, CLA, PHO, BCR, PL9, LMG, SQD, DGD, STE, BCT, LHG, HEM, HEC |
 |
Rt Xfel Structure Of Photosystem Ii 250 Microseconds After The Third Illumination At 2.09 Angstrom Resolution
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-03-22 Classification: PHOTOSYNTHESIS Ligands: OEY, OEX, FE2, CL, CLA, PHO, BCR, PL9, SQD, DGD, STE, LMG, LHG, BCT, HEM, HEC |
 |
Rt Xfel Structure Of Photosystem Ii 500 Microseconds After The Third Illumination At 2.03 Angstrom Resolution
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-03-22 Classification: PHOTOSYNTHESIS Ligands: OEY, FE2, CL, BCT, CLA, BCR, PL9, LMG, SQD, LHG, DGD, STE, PHO, HEM, HEC |
 |
Rt Xfel Structure Of Photosystem Ii 730 Microseconds After The Third Illumination At 2.03 Angstrom Resolution
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-03-22 Classification: PHOTOSYNTHESIS Ligands: OEY, OEX, FE2, CL, CLA, PHO, BCR, PL9, LMG, SQD, LHG, DGD, STE, BCT, HEM, HEC |

