Search Count: 31
 |
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (3 Picosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (300 Ps Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (1 Nanosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (3 Nanosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-11-22 Classification: LYASE Ligands: FDA, SO4 |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (10 Nanosecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-11-22 Classification: LYASE Ligands: FDA, SO4 |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (30 Nanosecond Timepoint)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (1 Microsecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (10 Microsecond Pump Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (30 Microsecond Pump-Probe Delay)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Time-Resolved Sfx Structure Of The Class Ii Photolyase Complexed With A Thymine Dimer (100 Microsecond Timpeoint)
Organism: Methanosarcina mazei go1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-11-22 Classification: LYASE Ligands: SO4, FDA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-09-14 Classification: HYDROLASE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-14 Classification: HYDROLASE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2022-09-14 Classification: HYDROLASE |
 |
Crystal Structure Of Nemo Cozi In Complex With Hoip Nzf1 And Linear Diubiquitin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2022-08-17 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-04-13 Classification: DNA BINDING PROTEIN Ligands: MG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-06 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-03-03 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-02-03 Classification: PROTEIN BINDING |
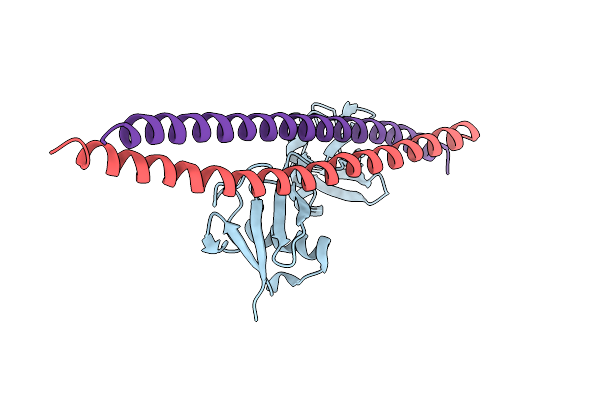 |
Crystal Structure Of Abin-1 Uban In Complex With One M1-Linked Di-Ubiquitin
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2019-07-17 Classification: SIGNALING PROTEIN |

