Search Count: 14
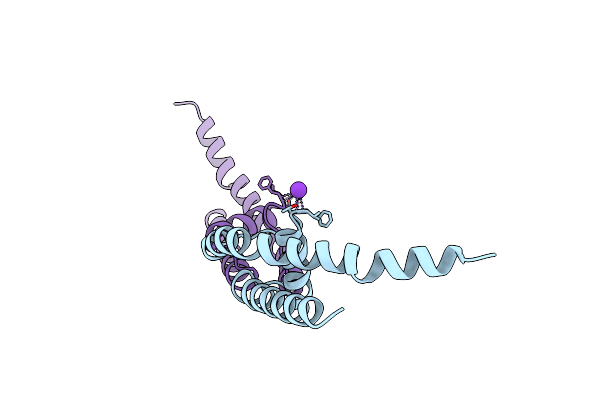 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-19 Classification: MEMBRANE PROTEIN Ligands: K |
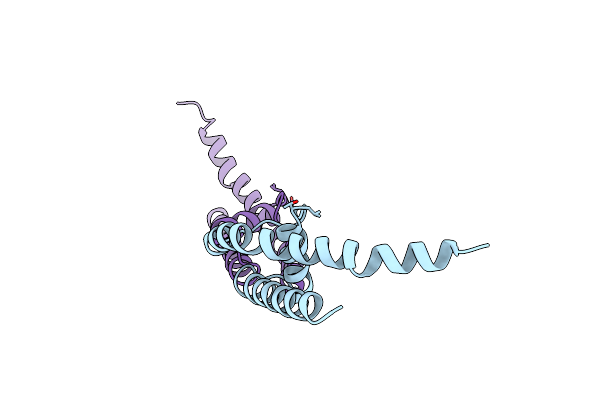 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-10-05 Classification: MEMBRANE PROTEIN Ligands: K |
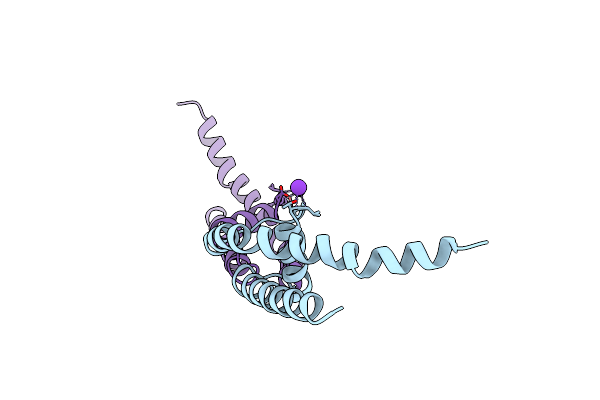 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-10-05 Classification: MEMBRANE PROTEIN Ligands: K |
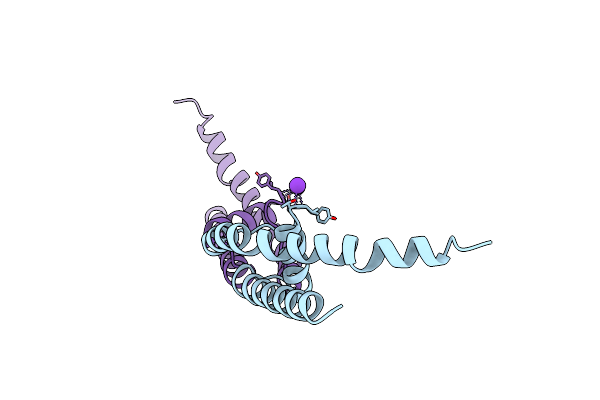 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-10-05 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-10-05 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-10-05 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2004-08-03 Classification: REPLICATION |
 |
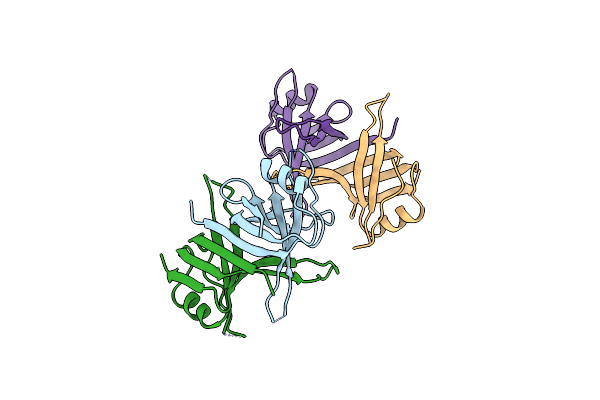
Crystal Structure Of Chymotryptic Fragment Of E. Coli Ssb Bound To Two 35-Mer Single Strand Dnas
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2000-08-01 Classification: replication/DNA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1997-12-31 Classification: DNA BINDING PROTEIN |
 |
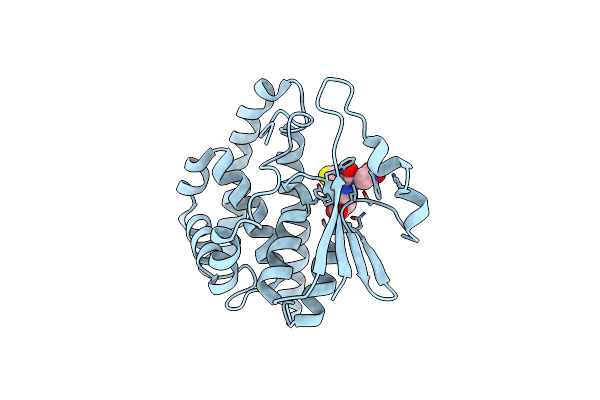
The Linker Of Des-Glu84 Calmodulin Is Bent As Seen In The Crystal Structure
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1994-05-31 Classification: CALCIUM-BINDING PROTEIN Ligands: CA |
 |
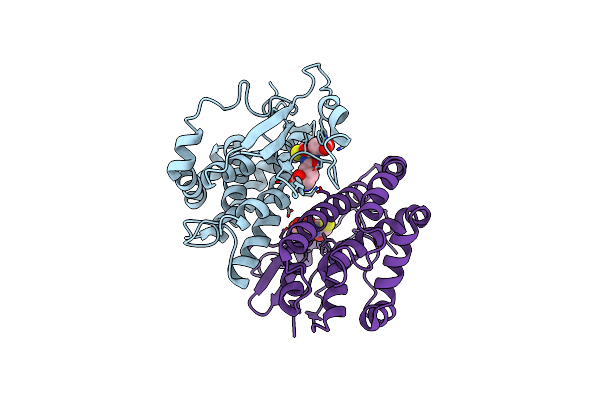
Crystal Structure Of Human Class Mu Glutathione Transferase Gstm2-2: Effects Of Lattice Packing On Conformational Heterogeneity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1994-01-31 Classification: TRANSFERASE(GLUTATHIONE) Ligands: GDN |
 |
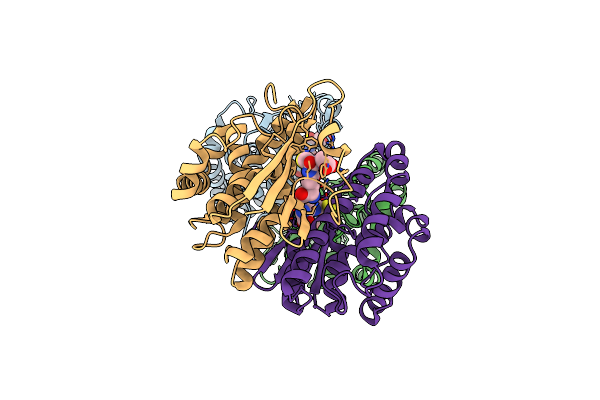
Crystal Structure Of Human Class Mu Glutathione Transferase Gstm2-2: Effects Of Lattice Packing On Conformational Heterogeneity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 1994-01-31 Classification: TRANSFERASE(GLUTATHIONE) Ligands: GDN |
 |
Crystal Structure Of Human Class Mu Glutathione Transferase Gstm2-2: Effects Of Lattice Packing On Conformational Heterogeneity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1994-01-31 Classification: TRANSFERASE(GLUTATHIONE) Ligands: GDN |
 |
Inhibition Of Proteinase K By Methoxysuccinyl-Ala-Ala-Pro-Ala-Chloromethyl Ketone. An X-Ray Study At 2.2-Angstroms Resolution
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |

