Search Count: 16
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-10-18 Classification: HYDROLASE |
 |
Structure Guided Design Of An Antibacterial Peptide That Targets Udp-N-Acetylglucosamine Acyltransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-01-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The Kdo Hydroxylase Kdoo, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase In Complex With Cobalt(Ii)
Organism: Methylacidiphilum infernorum (isolate v4)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-09-05 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, SO4, GOL, PG4, ACT, CL |
 |
Crystal Structure Of The Kdo Hydroxylase Kdoo, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase In Complex With Alphaketoglutarate And Fe(Iii)
Organism: Methylacidiphilum infernorum (isolate v4)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-09-05 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, AKG, GOL, PG4, ACT, SO4 |
 |
Crystal Structure Of The Kdo Hydroxylase Kdoo, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase In Complex With Succinate And Fe(Iii)
Organism: Methylacidiphilum infernorum
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-09-05 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, FE, GOL, SIN, ACT |
 |
Apo Structure Of The Kdo Hydroxylase Kdoo, A Non-Heme Fe(Ii) Alphaketoglutarate Dependent Dioxygenase
Organism: Methylacidiphilum infernorum (isolate v4)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2018-09-05 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT, SO4, PG4 |
 |
Organism: Caulobacter crescentus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: UDG |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-02-10 Classification: MEMBRANE PROTEIN Ligands: ZN, GOL, CXE |
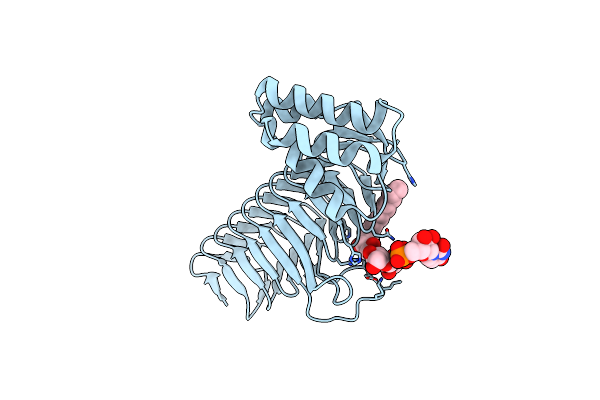 |
Structural Basis For The Acyl Chain Selectivity And Mechanism Of Udp-N-Acetylglucosamine Acyltransferase
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2007-10-02 Classification: TRANSFERASE Ligands: U20 |
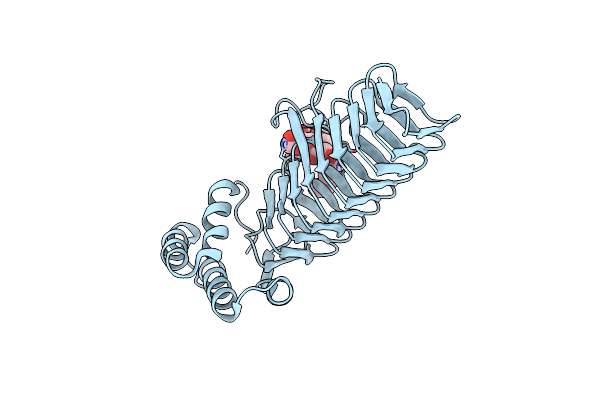 |
Structural Basis For The Acyl Chain Selectivity And Mechanism Of Udp-N-Acetylglucosamine Acyltransferase
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-10-02 Classification: TRANSFERASE Ligands: U21 |
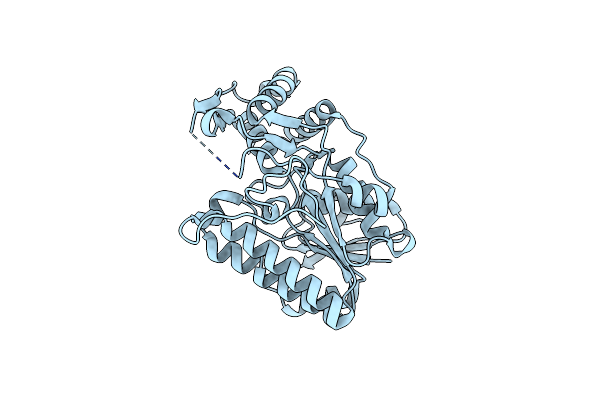 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-04-08 Classification: TRANSFERASE |
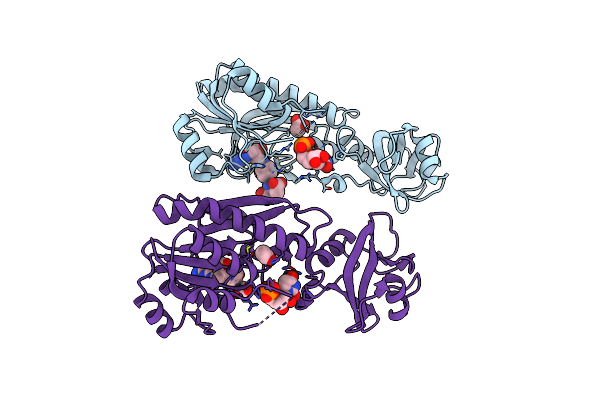 |
N-Terminal Formyltransferase Domain Of Arna In Complex With N-5- Formyltetrahydrofolate And Ump
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2005-04-08 Classification: TRANSFERASE Ligands: FON, U5P, ACT |
 |
Human Nuclear Receptor Liver Receptor Homologue-1, Lrh-1, Bound To Phospholipid And A Fragment Of Human Shp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-03-01 Classification: TRANSCRIPTION REGULATION Ligands: EPH, GOL |
 |
Organism: Aquifex aeolicus
Method: SOLUTION NMR Release Date: 2004-11-23 Classification: HYDROLASE Ligands: ZN, TUX |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2002-09-13 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2002-09-13 Classification: TRANSFERASE |

