Search Count: 20
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: ZN, CA, CL, EPE |
 |
Crystal Structure Of Ndm-12 Metallo-Beta-Lactamase In Complex With Hydrolyzed Ampicillin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-22 Classification: hydrolase/antibiotic Ligands: ZN, CL, NA, ZZ7 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: ZN, CL, PO4, EDO, PEG, PGE, GOL |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-08-28 Classification: HYDROLASE Ligands: SO4, ETX, GOL, CL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-04-10 Classification: DNA BINDING PROTEIN Ligands: FLC, ADP, EDO, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-04-10 Classification: DNA BINDING PROTEIN/DNA Ligands: EDO, PGE, ZN, FLC |
 |
Crystal Structure Of Ndm-1 In Complex With Hydrolyzed Cefuroxime - New Refinement
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ZN, SO4, 3S0, UNL |
 |
Crystal Structure Of Ndm-1 In Complex With Hydrolyzed Ampicillin - New Refinement
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2018-12-26 Classification: HYDROLASE Ligands: ZN, ZZ7, SO4, CL, EDO |
 |
Ndm-1 Metallo-Beta-Lactamase: A Parsimonious Interpretation Of The Diffraction Data
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-10-03 Classification: HYDROLASE Ligands: ZN, PG4, CL, P6G |
 |
Crystal Structure Of Ndm-1 Metallo-Beta-Lactamase In Complex With Cd Ions And A Hydrolyzed Beta-Lactam Ligand - New Refinement
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: CD, CL, UNL, PG4, TOE, EDO |
 |
Bacillus Cereus Zn-Dependent Metallo-Beta-Lactamase At Ph 7 - New Refinement
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-07-12 Classification: HYDROLASE Ligands: ZN, UNX, SO4 |
 |
Crystal Structure Of Ndm-1 In Complex With Hydrolyzed Meropenem - New Refinement
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-04-05 Classification: HYDROLASE/ANTIBIOTIC Ligands: 8YL, ZN, GOL, SO4 |
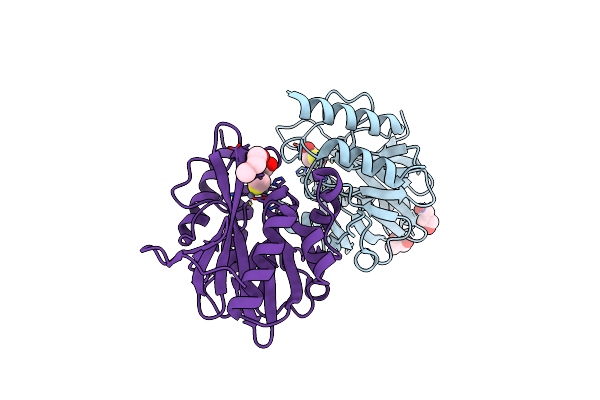 |
Crystal Structure Of Ndm-1 In Complex With Beta-Mercaptoethanol - New Refinement
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2017-04-05 Classification: HYDROLASE Ligands: ZN, BME, GOL, CL, PG4 |
 |
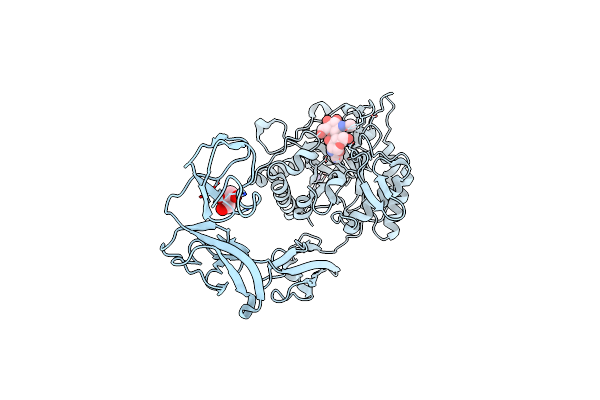
Organism: Vibrio marinus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
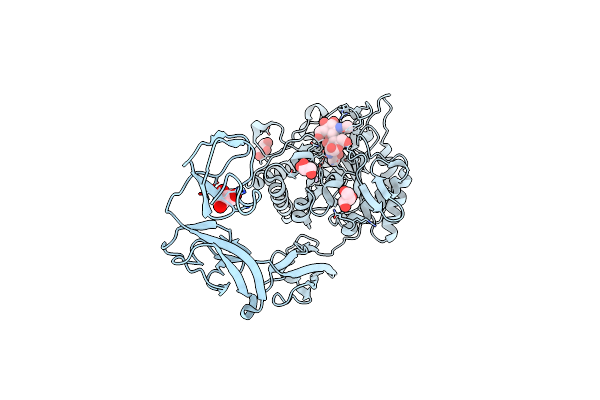
Crystal Structure Of Cold-Adapted Chitinase From Moritella Marina With A Reaction Intermediate - Oxazolinium Ion (Ngo)
Organism: Vibrio marinus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2013-05-01 Classification: HYDROLASE Ligands: NAG, NGO, NA, GOL |
 |
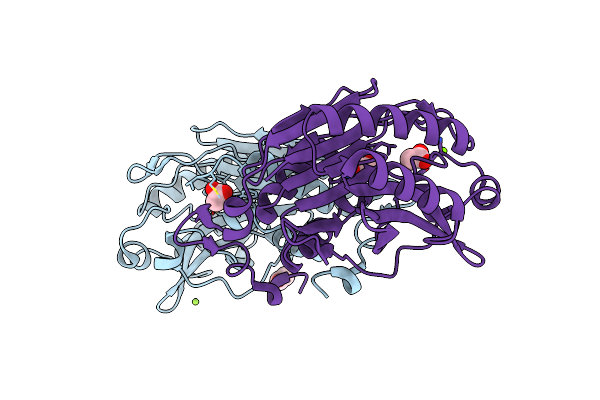
Crystal Structure Of Cold-Adapted Chitinase From Moritella Marina With A Reaction Product - Nag2
Organism: Vibrio marinus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-11-24 Classification: HYDROLASE Ligands: ACY, MG |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-11-24 Classification: HYDROLASE Ligands: BR, ACY, MG |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2010-09-22 Classification: HYDROLASE Ligands: MG |
 |
Organism: Pyrococcus abyssi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-22 Classification: HYDROLASE Ligands: ACY, MG |

