Search Count: 56
 |
Organism: Acinetobacter silvestris
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL ADHESION Ligands: ACT |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Micromonospora maris ab-18-032
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-09-18 Classification: LIGASE Ligands: EPE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-09 Classification: ELECTRON TRANSPORT Ligands: HEM, LMR, CL |
 |
Organism: Streptomyces sp. nl15-2k
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-06-21 Classification: LIGASE Ligands: IMD, PEG, EDO, GLY, SER, GOL |
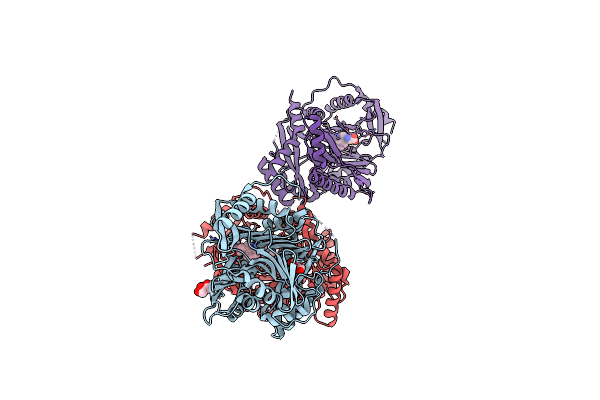 |
Structure Of Aminodeoxychorismate Synthase Component 1 (Pabb) From Bacillus Subtilis Spizizenii.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: GOL, TRP |
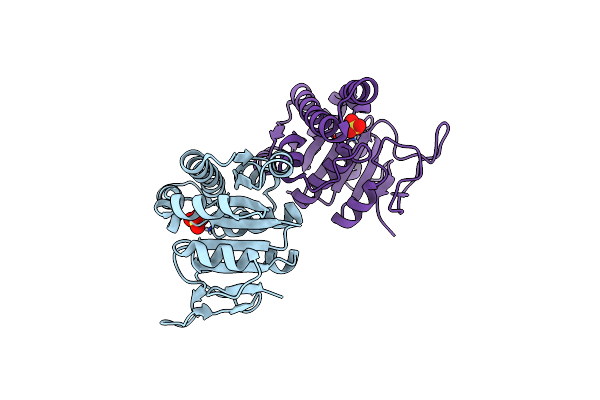 |
Crystal Structure Of The 5-(Aminomethyl)Furan-3-Yl Methyl Phosphate Kinase Mfne From Methanococcus Vannielii.
Organism: Methanococcus vannielii
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: SO4 |
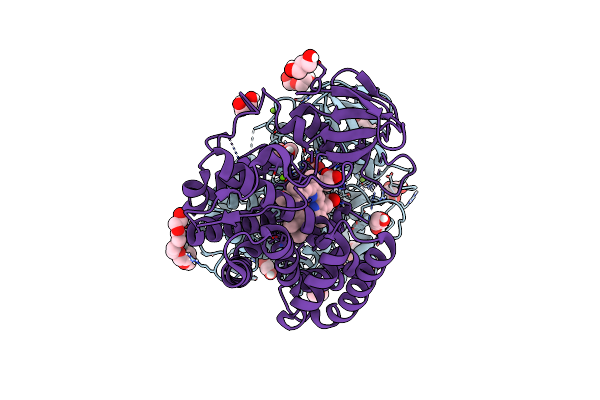 |
Organism: Micromonospora maris (strain dsm 45365 / jcm 31040 / nbrc 109089 / nrrl b-24793 / ab-18-032)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-11-30 Classification: OXIDOREDUCTASE Ligands: HEM, PGE, GOL, PEG, P6G, P33, CL, MG |
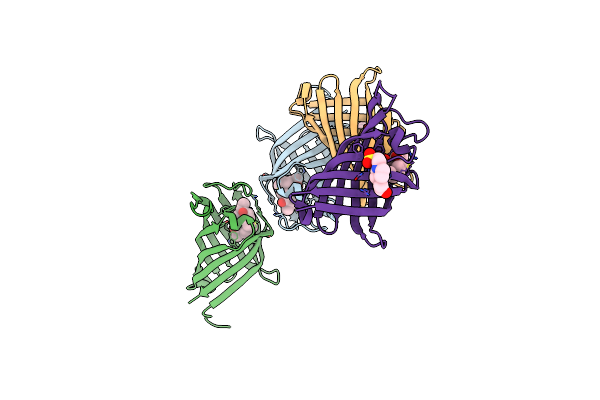 |
Structure Of The Diels Alderase Enzyme Abyu, From Micromonospora Maris, Co-Crystallised With A Non Transformable Substrate Analogue
Organism: Micromonospora maris ab-18-032
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-16 Classification: LIGASE Ligands: 8IF, EPE |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: PG4, BOG, NA |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-09-07 Classification: TRANSFERASE Ligands: MG, TRP |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-10-06 Classification: DE NOVO PROTEIN Ligands: HEM |
 |
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-05-26 Classification: CELL ADHESION |
 |
Organism: Streptomyces koyangensis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-04-21 Classification: LIGASE Ligands: BR |
 |
Nmr Structure Of Repeat Domain 13 Of The Fibrillar Adhesin Csha From Streptococcus Gordonii.
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: SOLUTION NMR Release Date: 2020-04-08 Classification: CELL ADHESION |
 |
Solution Structure Of Macpd, A Acyl Carrier Protein, From Pseudomonas Fluorescens Involved In Mupirocin Biosynthesis.
Organism: Pseudomonas fluorescens
Method: SOLUTION NMR Release Date: 2020-02-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Verrucosispora
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-05-16 Classification: HYDROLASE |
 |
Crystal Structure Of 2-Methylcitrate Dehydratase (Mmge) From Bacillus Subtilis.
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-02-14 Classification: LYASE Ligands: TLA |
 |
Crystal Structure Of 2-Methylcitrate Dehydratase (Prpd) From Salmonella Enterica
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2018-02-14 Classification: LYASE |

