Search Count: 71,744
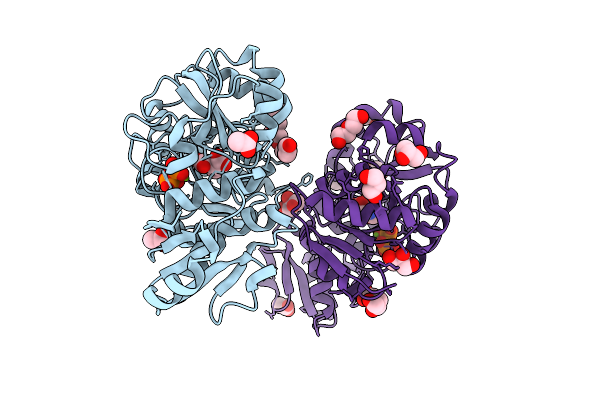 |
Thermotoga Maritima Threonylcarbamoyl Adenylate Synthase (Tsac2) In Complex With N-Carboxy-L-Threonine, Magnesium And Pyrophosphate.
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: U6A, MG, DPO, P6G, ACT, GOL, PEG |
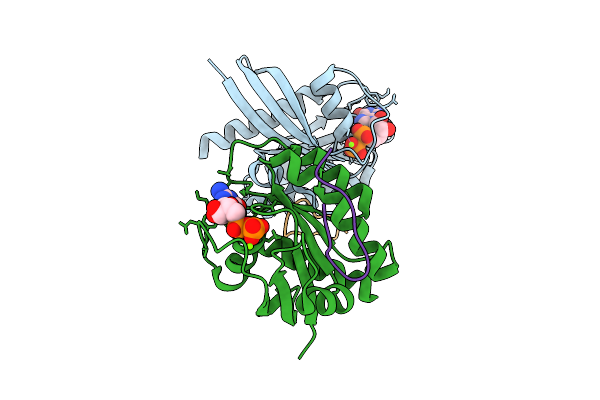 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GDP |
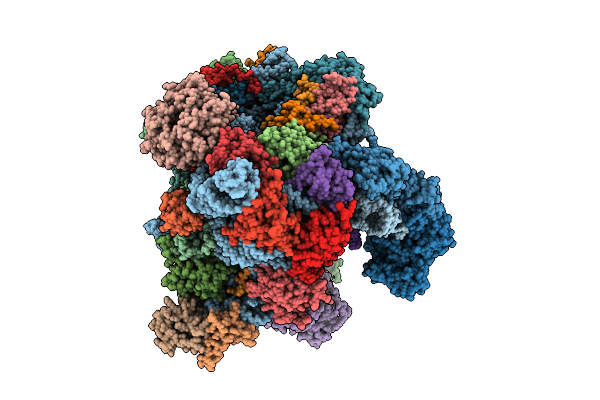 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
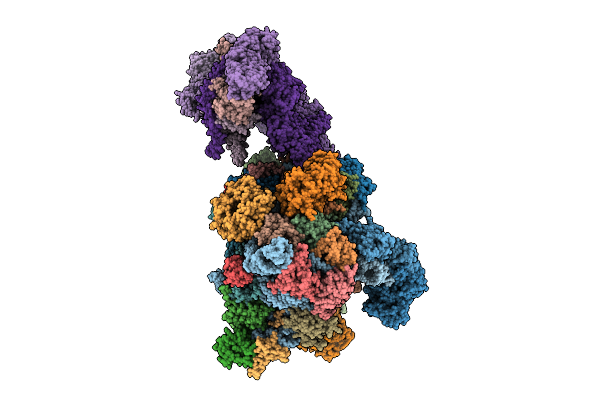 |
Late-Stage 48S Initiation Complex With Eif3 (Ls48S-Eif3 Ic) Guided By The Trans-Rna
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING Ligands: A1EHH |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: A1L6U, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: NAG, A1L6V |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: NAG, A1L6W |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: NAG, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: A1L6U |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: NAG, A1L6V |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: NAG, A1L6W |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: NAG |
 |
Organism: Thermus phage philo
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ANTIVIRAL PROTEIN/DNA/RNA Ligands: ATP, ZN, MG |
 |
High-Resolution Crystal Structure Of Methyl-2,3-Diamino Propanoic Acid-Ams Inhibitor Bound Adenylation Domain (A3) From Sulfazecin Nonribosomal Peptide Synthetase Sulm
Organism: Paraburkholderia acidicola
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: A1BVA, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Homo sapiens, Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ONCOPROTEIN Ligands: GDP, MG, EDO, A1BV7 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA Ligands: SO4, MG |

