Search Count: 24
 |
Organism: Rhodospirillum rubrum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: S5Q, CLF, ADP, MG, AF3, SF4 |
 |
Organism: Rhodobacter capsulatus sb 1003
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: OXIDOREDUCTASE Ligands: S5Q, HCA, CLF, ADP, MG, AF3, SF4 |
 |
Chapso Treated Partial Catalytic Component (Comprising Only Anfd & Anfk, Lacking Anfg And Fefeco) Of Iron Nitrogenase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus sb 1003
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: OXIDOREDUCTASE Ligands: CLF |
 |
Organism: Rhodobacter capsulatus sb 1003
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-01-19 Classification: RNA BINDING PROTEIN Ligands: EDO, BME |
 |
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: FES, HEC |
 |
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSLOCASE/Oxidoreductase Ligands: HEC, FES |
 |
R. Capsulatus Cyt Bc1 With One Fes Protein In B Position And One In C Position (Ciii2 B-C)
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSLOCASE/Oxidoreductase Ligands: FES, HEC |
 |
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSLOCASE/Oxidoreductase Ligands: HEC, FES |
 |
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSLOCASE/Oxidoreductase Ligands: HEC, CU, FES |
 |
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSLOCASE/Oxidoreductase Ligands: FES, HEC, CU |
 |
Organism: Rhodobacter capsulatus (strain atcc baa-309 / nbrc 16581 / sb1003)
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSLOCASE/Oxidoreductase Ligands: FES, HEC, CU |
 |
Cryo-Em Structure Of Nadh Reduced Form Of Nad+-Dependent Formate Dehydrogenase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Resolution:3.24 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: MGD, 6MO, FES, SF4, H2S, FMN, NAI |
 |
Cryo-Em Structure Of As Isolated Form Of Nad+-Dependent Formate Dehydrogenase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2020-04-22 Classification: OXIDOREDUCTASE Ligands: MGD, 6MO, FES, SF4, H2S, FMN |
 |
Organism: Starkeya novella
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2011-05-18 Classification: Heme-binding protein/Heme-binding protein Ligands: HEC, SO4 |
 |
Organism: Starkeya novella
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-05-18 Classification: Heme-binding protein Ligands: HEC |
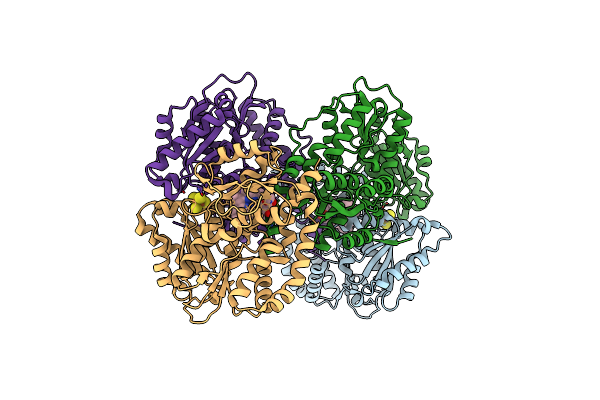 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
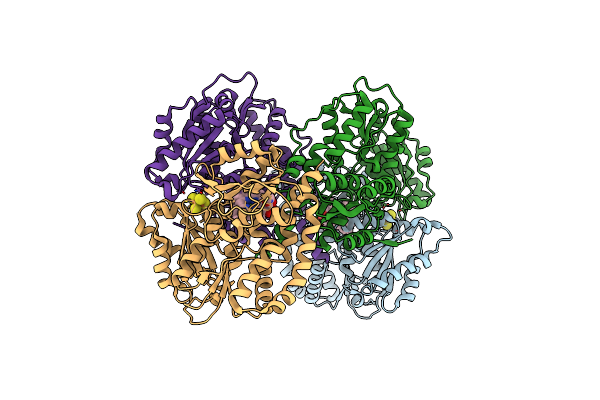 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
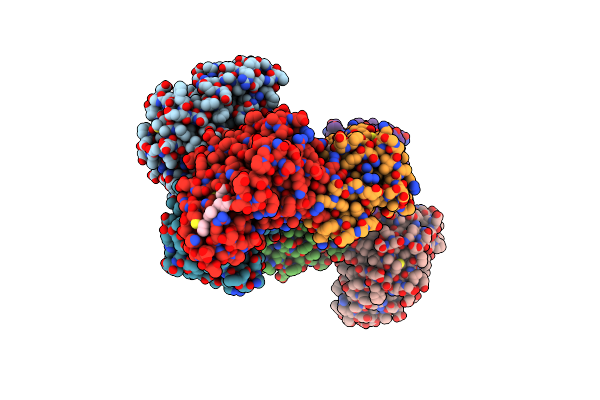 |
Organism: Rhodovulum sulfidophilum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-01-08 Classification: ELECTRON TRANSPORT Ligands: HEC |
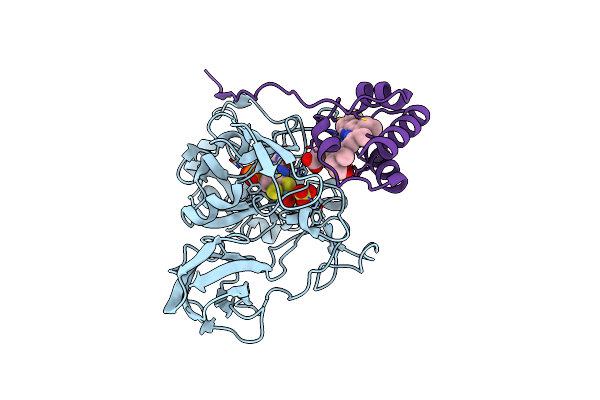 |
Organism: Thiobacillus novellus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-02-20 Classification: OXIDOREDUCTASE Ligands: MSS, SO4, HEC |
 |
Organism: Starkeya novella
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-02-20 Classification: OXIDOREDUCTASE Ligands: MSS, HEC |

