Search Count: 538
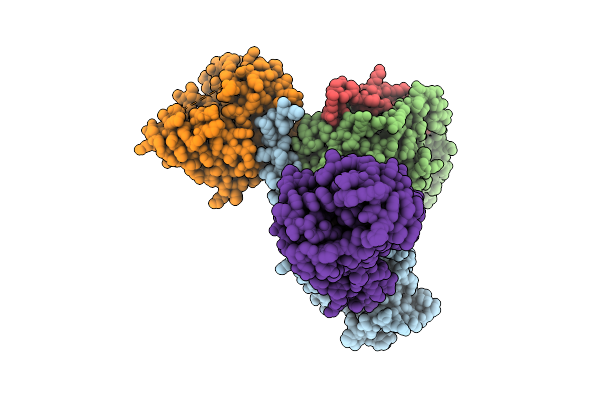 |
Cryo-Em Structure Of Active Human Green Cone Opsin In Complex With Chimeric G Protein (Minigist)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: SIGNALING PROTEIN Ligands: RET |
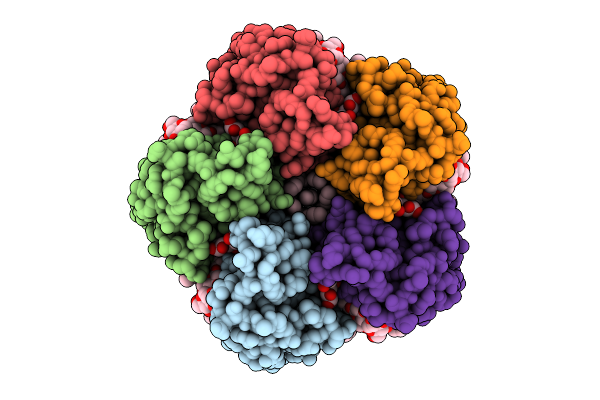 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
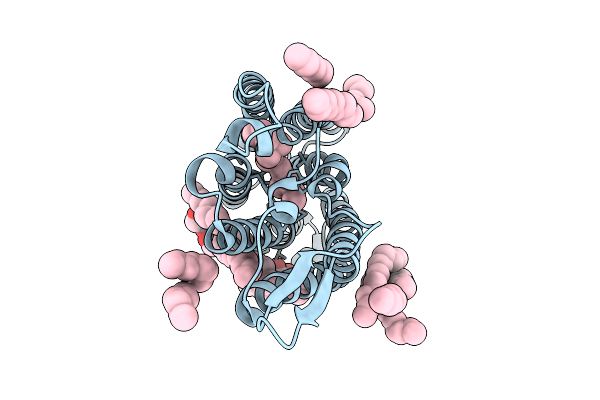 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The K2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
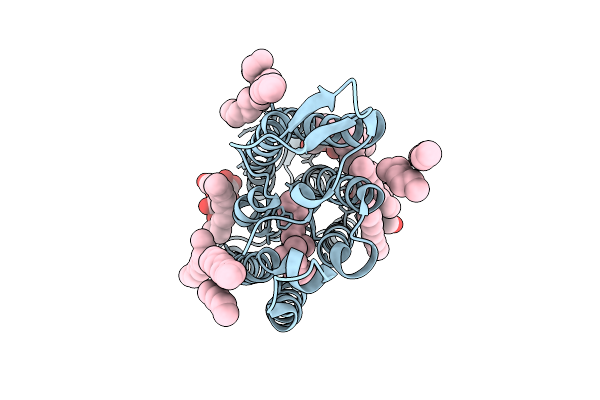 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The O2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: RET, 3PE, D12 |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: HEX, D12, RET |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Room Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 95% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Room-Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 85% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Organism: Klebsormidium nitens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ELECTRON TRANSPORT Ligands: PC1, RET |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: STRUCTURAL PROTEIN Ligands: RET |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: DE NOVO PROTEIN Ligands: RET, SO4 |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |

