Search Count: 70,499
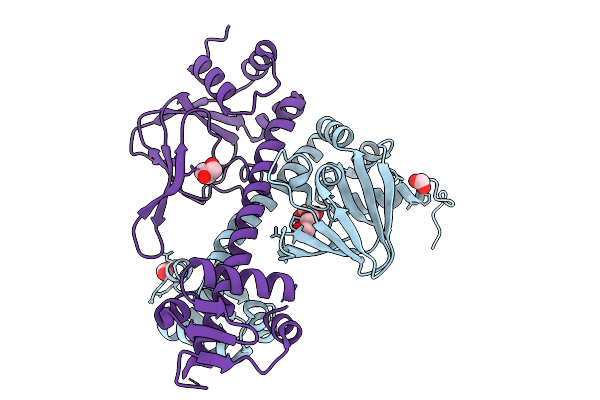 |
Crystal Structure Of The Transcriptional Regulator Hcpr From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-28 Classification: TRANSCRIPTION Ligands: GOL, FMT |
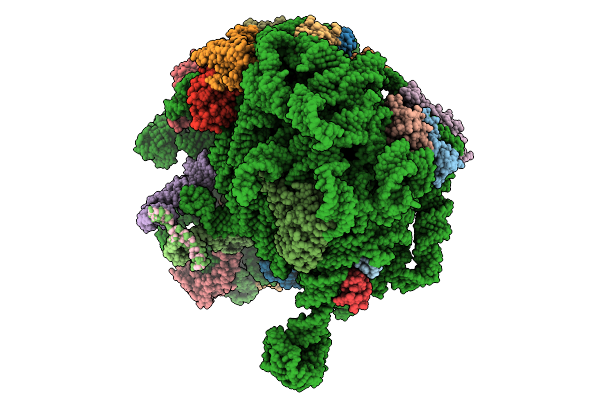 |
Structure Of E.Coli Ribosome With Nascent Chain At Linker Length Of 31 Amino Acids, With Mrna, P-Site And A-Site Trnas
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME |
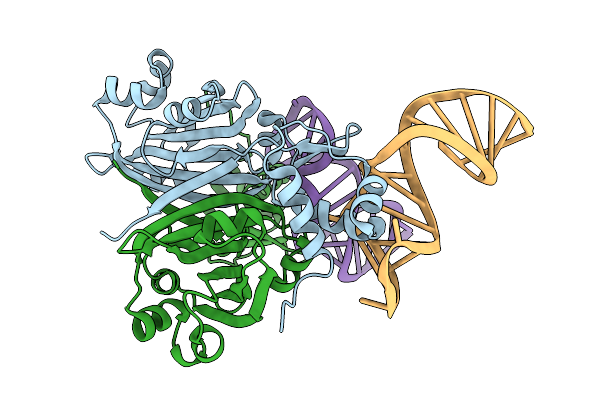 |
Crystal Structure Of Dasr In Complex With A Synthetic Dasr-Binding Rna Aptamer
Organism: Streptomyces coelicolor, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: BR |
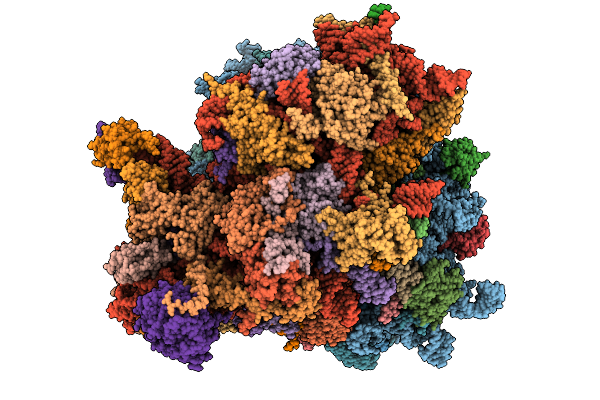 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME Ligands: K, MG, NA, SPD, PUT, ZN, HYG |
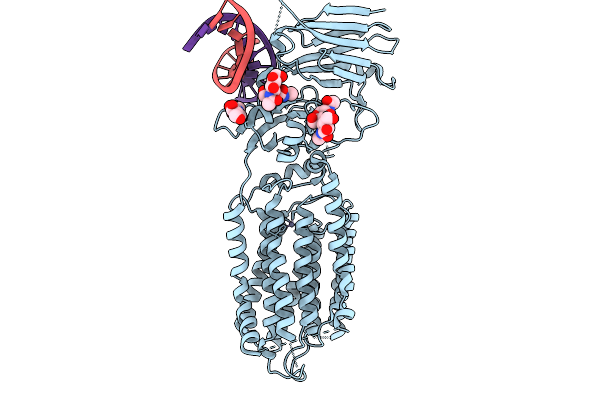 |
Organism: Caenorhabditis elegans, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.99 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/RNA Ligands: NAG, ZN |
 |
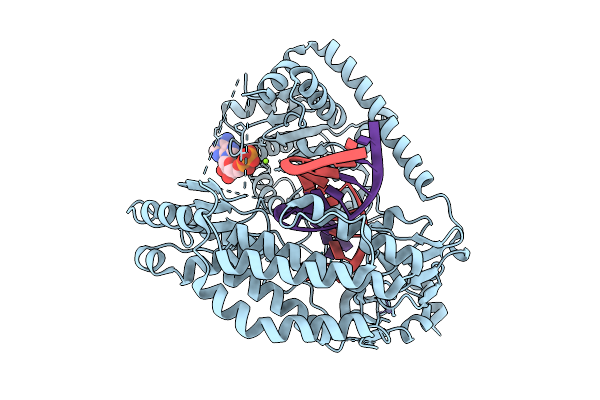
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: A1EY1, ECC |
 |
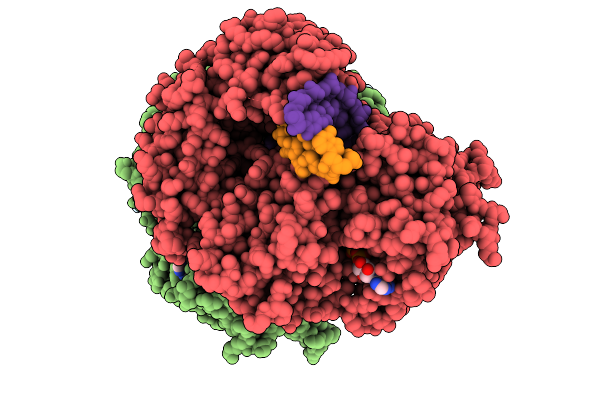
Organism: Homo sapiens, Uncultured archaeon
Method: ELECTRON MICROSCOPY Resolution:3.36 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM/RNA Ligands: ADP, ZN, MG |
 |
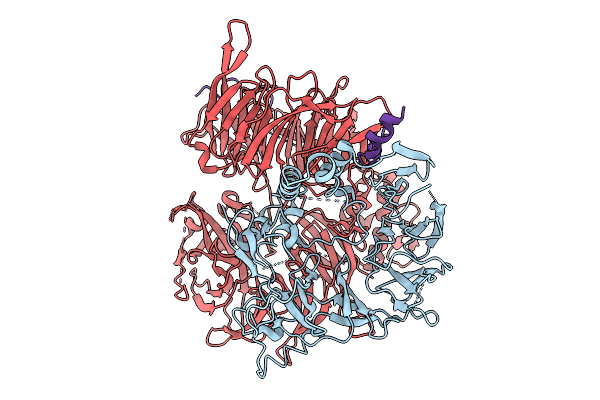
Organism: Homo sapiens, Uncultured archaeon
Method: ELECTRON MICROSCOPY Resolution:3.07 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM/RNA Ligands: ADP, ZN, MG |
 |
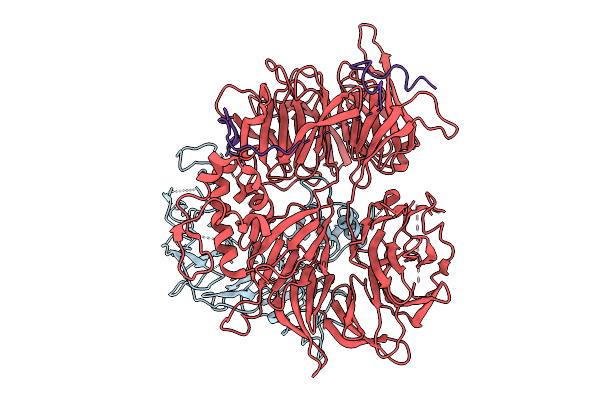
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.65 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
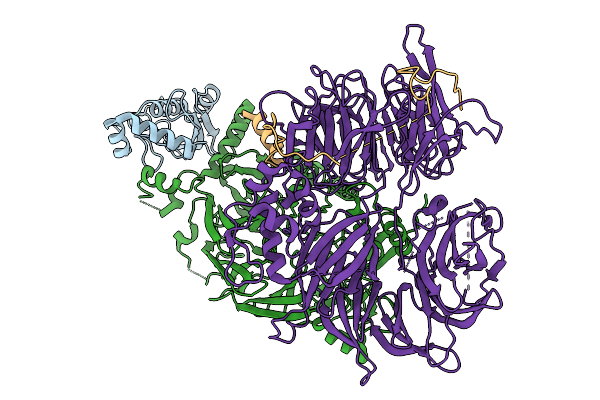
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.93 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
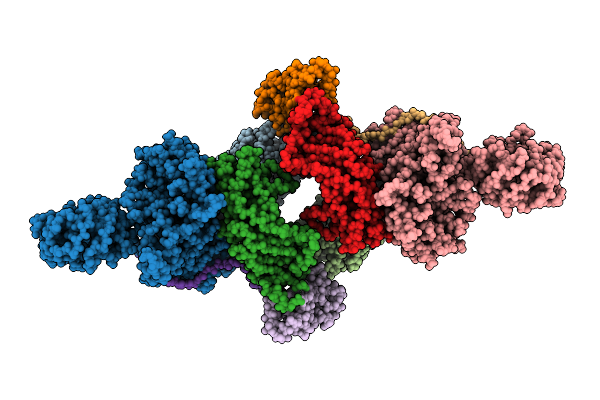
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
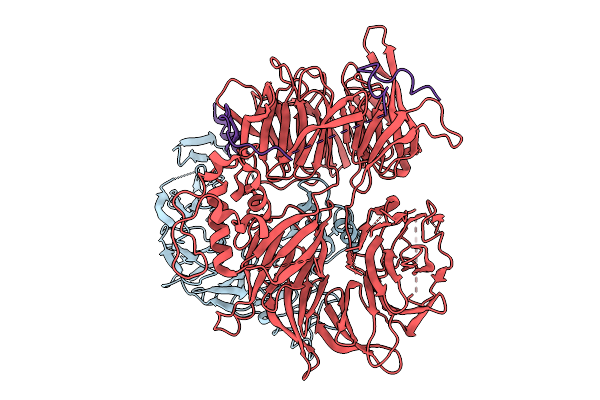
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.03 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.25 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.62 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
Local Refinement Of Stacked Like Ddb1-Dda1-Det1-Ube2E2-Cop1 Complex (Layer 1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.99 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
Photo-Crosslinkable Oligonucleotodes Which Have A Trioxsalen Conjugated To C2' Position Of Guanosine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2026-01-28 Classification: DNA Ligands: A1L7U |
 |
Local Refinement Of Stacked Like Ddb1-Dda1-Det1-Ube2E2-Cop1 Complex (Layer 2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.25 Å Release Date: 2026-01-28 Classification: LIGASE |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN |
 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |

