Search Count: 71,339
 |
Crystal Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3 In Complex With Rgg3Bp13
Organism: Streptococcus thermophilus lmg 18311, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2026-02-04 Classification: DNA BINDING PROTEIN Ligands: SO4, GOL, TRS |
 |
Crystal Structure Of Sorghum Sulfotransferase Lgs1 Reveals Sulfation-Assisted Bc-Ring Formation In Strigolactone Biosynthesis
Organism: Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: A3P, GOL |
 |
Organism: Psychrobacter lutiphocae dsm 21542, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN/RNA Ligands: CA, GOL, PEG |
 |
Tubulin Cofactors D,E,G,C And Tubulin Complex -- Tbcc N Terminus Bound To Tubulin
Organism: Saccharomyces cerevisiae, Escherichia coli, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: GTP, GDP |
 |
Tubulin Cofactors D,E,G,C Bound To Tubulin Dimer -- Tbcc N Terminus Unbound
Organism: Saccharomyces cerevisiae, Escherichia coli, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: GTP, GDP |
 |
Organism: Saccharomyces cerevisiae, Escherichia coli, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: GTP, GDP |
 |
Organism: Saccharomyces cerevisiae, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: GTP, GDP |
 |
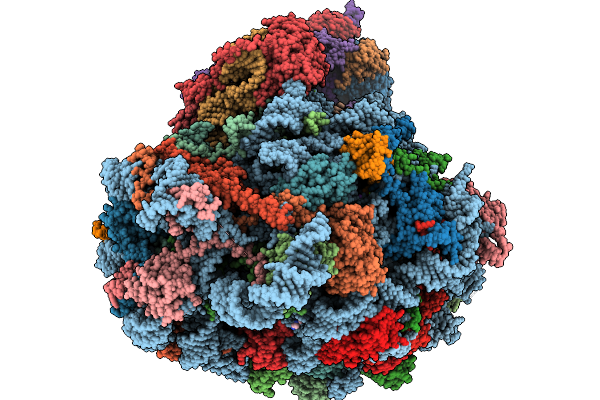
Structure Of The Arabidopsis Thaliana 80S Ribosome In Complex With P- And E-Site Trnas And Mrna
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:1.82 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, K, TER, SPD, EPE, ZN |
 |
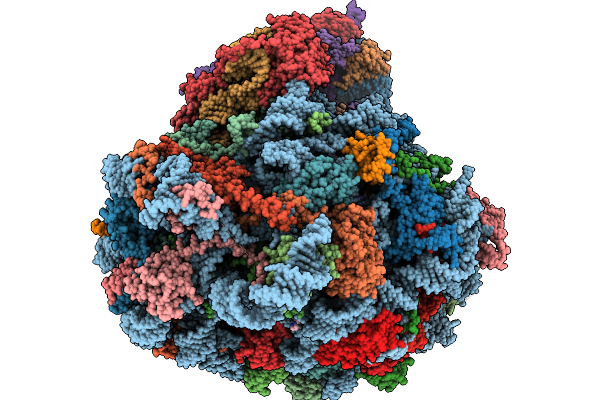
Structure Of The Arabidopsis Thaliana 80S Ribosome In Complex With P- And E-Site Trnas, Mrna, And Thermospermine
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: TER, MG, K, SPD, EPE, ZN |
 |
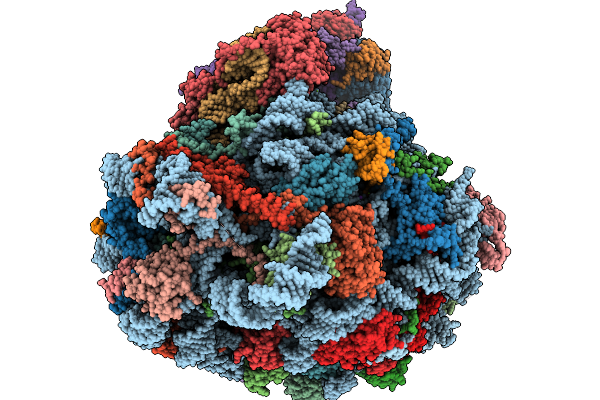
Structure Of The Arabidopsis Thaliana 80S Ribosome Ovac Mutant In Complex With P- And E-Site Trnas, Mrna, And Thermospermine
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.25 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: TER, MG, K, SPD, EPE, ZN |
 |
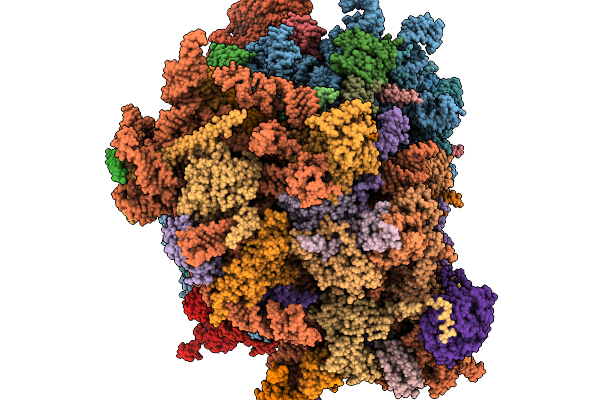
Structure Of The Arabidopsis Thaliana 80S Ribosome Ovac Mutant In Complex With P- And E-Site Trnas And Mrna
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.25 Å Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, K, TER, SPD, EPE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Escherichia coli nctc 86
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Escherichia coli nctc 86
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2026-02-04 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-04-154 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2026-02-04 Classification: ANTIMICROBIAL PROTEIN Ligands: A1I1F, A1IZN |
 |
Structure Of E.Coli Ribosome With Filamin Mutant Y719E Nascent Chain At Linker Length Of 47 Amino Acids, With Trna
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-04-161 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2026-02-04 Classification: ANTIMICROBIAL PROTEIN Ligands: A1I0A, A1I0B, EDO |
 |
Structure Of Carbamoylated Recombinant Human Butyrylcholinesterase By The Biscarbamte 5-(1-Hydroxy-2-(Piperidin-1-Yl)Ethyl)-1,3-Phenylene Bis(Piperidine-1-Carboxylate)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2026-02-04 Classification: HYDROLASE Ligands: NAG, MES, GOL, A1I0D, A1I0F, SO4 |
 |
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN/RNA |

