Search Count: 50
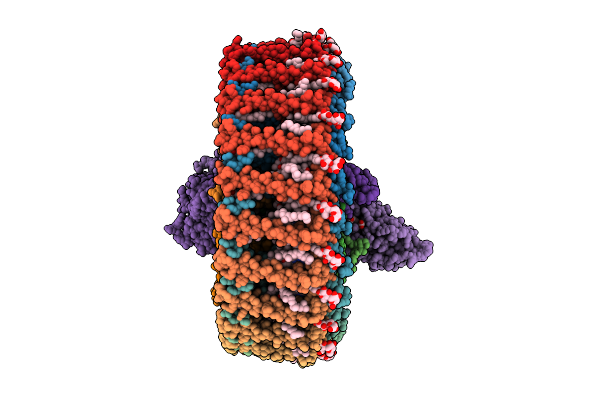 |
Cryo Em Structure Of Rc-Dlh Complex Model Ii From Gemmatimonas Groenlandica
Organism: Gemmatimonas groenlandica
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, LMT, MQ8, FE, CD4, CRT, PEX, HEC, V7N |
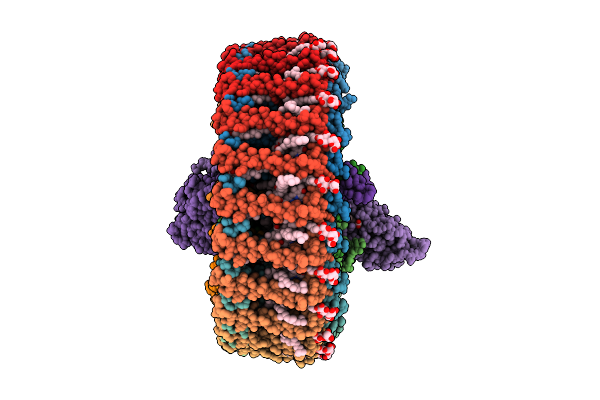 |
Cryo-Em Structure Of Rc-Dlh Complex Model I From Gem. Groenlandica Strain Tet16
Organism: Gemmatimonas groenlandica
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PHOTOSYNTHESIS Ligands: V7N, BCL, LMT, PEX, MQ8, CD4, BPH, FE, CRT, HEC |
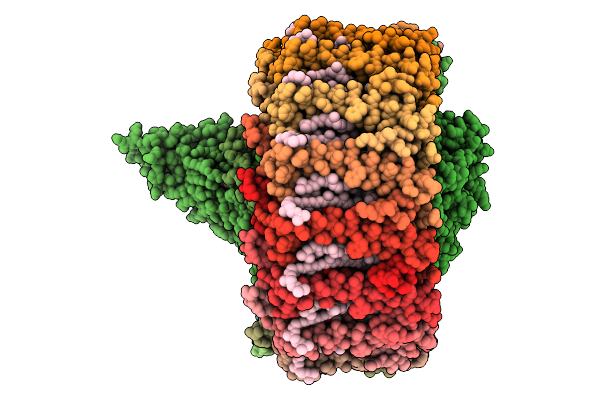 |
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PHOTOSYNTHESIS Ligands: U10, BCL, SPN, MW9, HEM, BPH, FE |
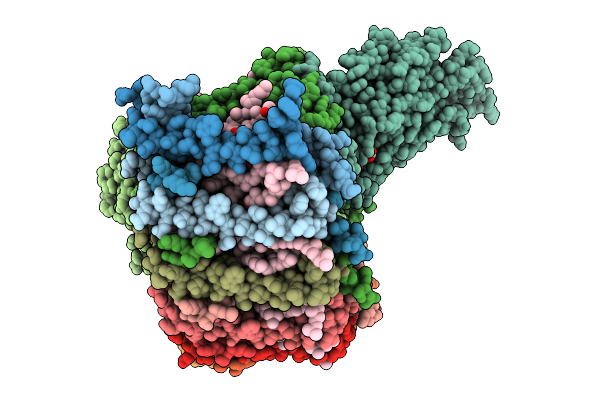 |
Cryo-Em Structure Of The Dinoroseobacter Shibae Rc-Lh1 Supercomplex With Incomplete Lh1 Ring(State 1)
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PHOTOSYNTHESIS Ligands: BCL, SPN, HEM, U10, BPH, FE |
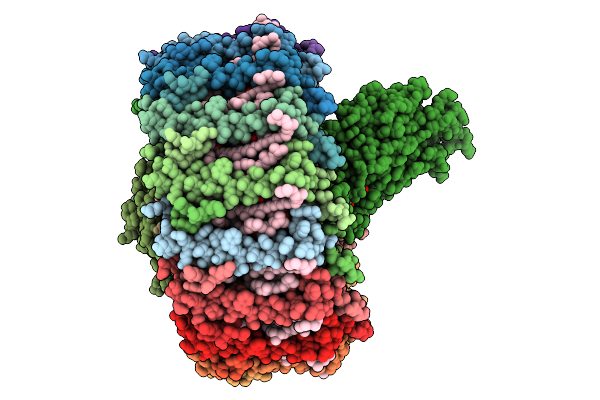 |
Cryo-Em Structure Of The Dinoroseobacter Shibae Rc-Lh1 Supercomplex With Incomplete Lh1 Ring(State 2)
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PHOTOSYNTHESIS Ligands: BCL, SPN, HEM, U10, BPH, FE |
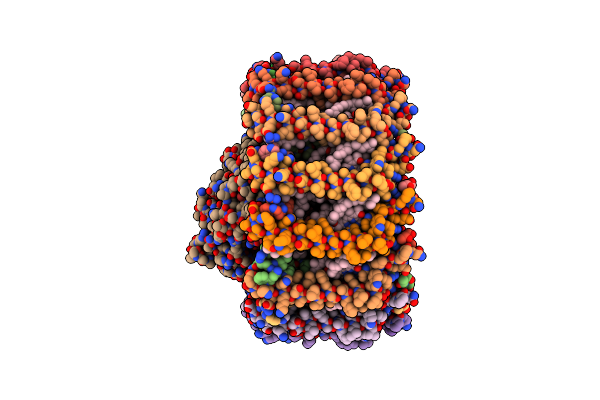 |
Organism: Rhodospirillum rubrum
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: PHOTOSYNTHESIS Ligands: 07D, CRT, FE, BPH, U10 |
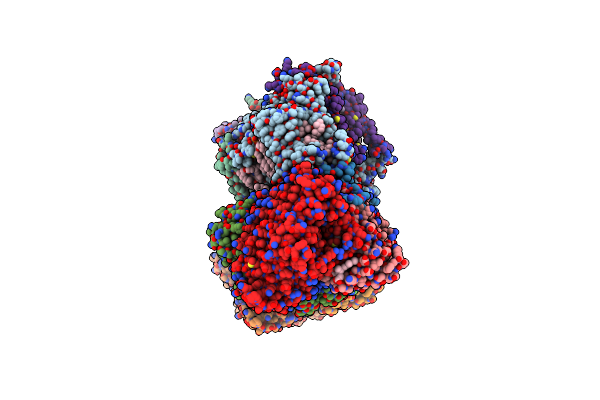 |
Cryo-Em Structure Of The Whole Photosynthetic Complex From The Green Sulfur Bacteria
Organism: Chlorobaculum tepidum tls
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: PHOTOSYNTHESIS Ligands: GS0, CLA, BCL, F39, LHG, IKV, CA, SF4 |
 |
Lipidic Cubic Phase Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Centre
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, HTO, OLC, SO4, BCB, BPB, UQ1, LDA, FE2, MQ7, NS5 |
 |
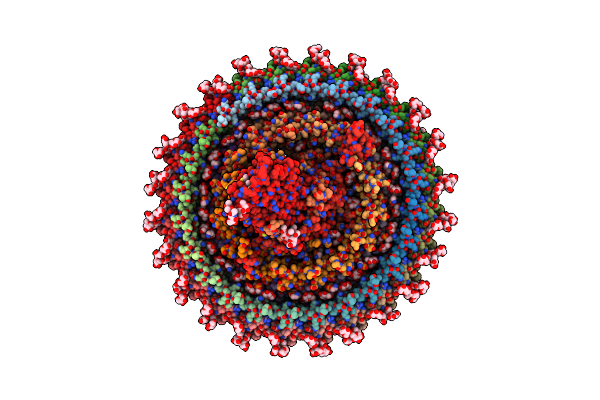
Lipidic Cubic Phase Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Centre
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, LDA, SO4, BCB, BPB, HTO, UQ2, FE2, MQ9, NS5, OLC |
 |
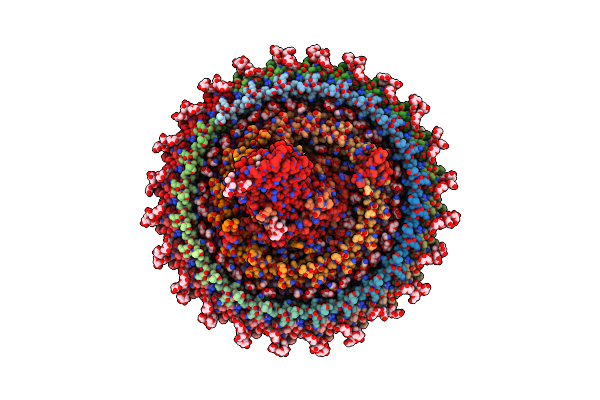
Cryo-Em Structure (Model_1A) Of The Rc-Dlh Complex From Gemmatimonas Phototrophica At 2.4 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, 0V9, HEC, NDG, V75, CD4, PGW, MQ8, BPH, FE, CRT, V7B, UYH |
 |
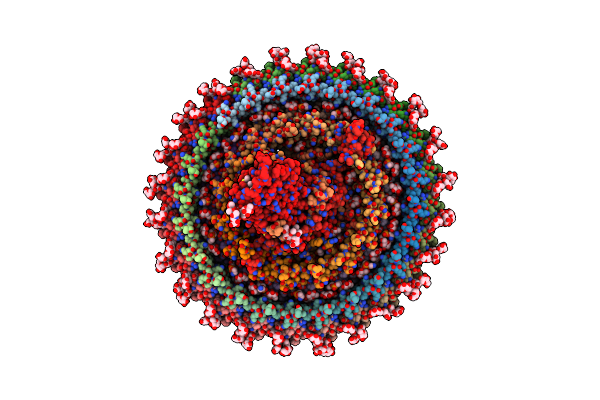
Cryo-Em Structure (Model_2A) Of The Rc-Dlh Complex From Gemmatimonas Phototrophica At 2.5 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, HEC, V75, NDG, 0V9, CD4, PGW, MQ8, V7B, BPH, FE, CRT, UYH |
 |
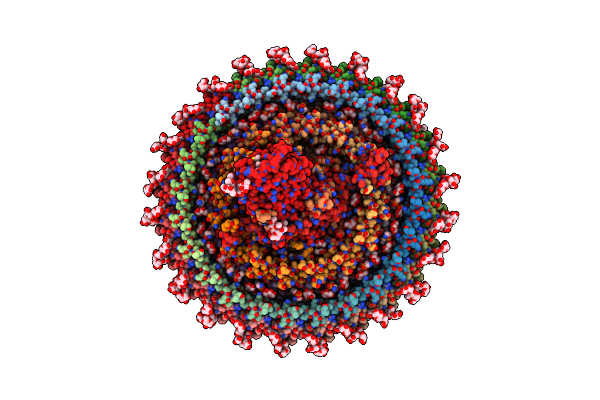
Cryo-Em Structure Of The Rc-Dlh Complex (Model_1B) From Gemmatimonas Phototrophica At 2.47 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, 0V9, HEC, V75, NDG, PGW, CD4, BPH, MQ8, FE, CRT, V7B, UYH |
 |
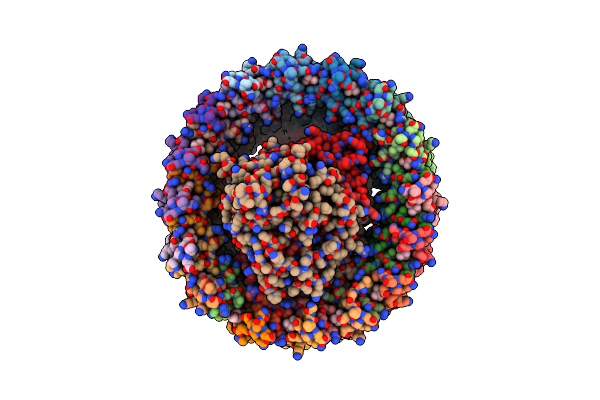
Cryo-Em Structure (Model_2B) Of The Rc-Dlh Complex From Gemmatimonas Phototrophica At 2.44 A
Organism: Gemmatimonas phototrophica
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: MEMBRANE PROTEIN Ligands: BCL, LMT, V7N, HEC, NDG, V75, PGW, 0V9, CD4, BPH, MQ8, FE, CRT, V7B, UYH |
 |
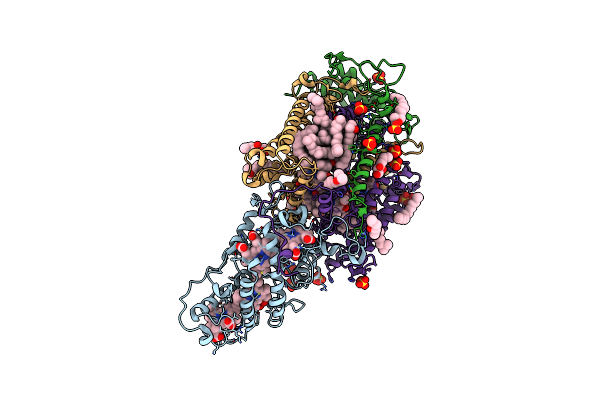
Organism: Rhodospirillum rubrum (strain atcc 11170 / ath 1.1.1 / dsm 467 / lmg 4362 / ncimb 8255 / s1)
Method: ELECTRON MICROSCOPY Release Date: 2021-09-22 Classification: STRUCTURAL PROTEIN Ligands: 07D, CRT, PGW, CD4, BPH, RQ0, U10, FE, CL, PO4, LMT |
 |
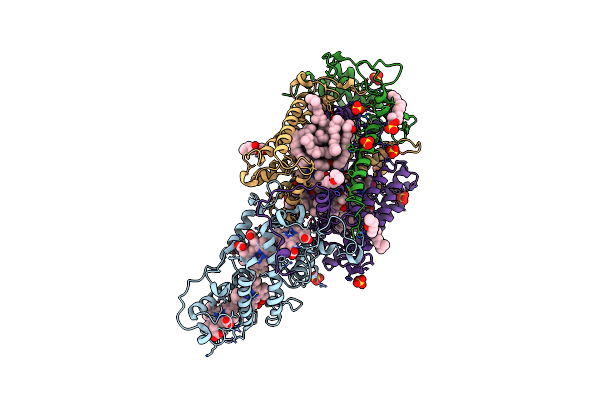
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
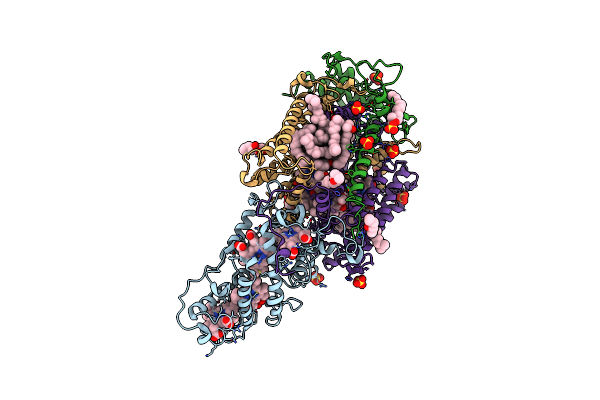
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
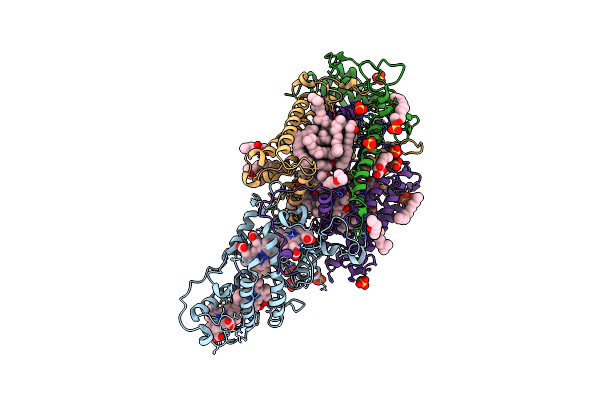
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |

