Search Count: 6
 |
Crystal Structure Of C. Difficile Penicillin-Binding Protein 2 In Complex With Ceftobiprole
Organism: Clostridioides difficile (strain r20291)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-03-23 Classification: PEPTIDE BINDING PROTEIN Ligands: RB6, ZN |
 |
Crystal Structure Of Pseudomonas Aeruginosa Penicillin-Binding Protein 3 (Pbp3) Complexed With Ceftobiprole
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-03-11 Classification: HYDROLASE Ligands: RB6, CL |
 |
Crystal Structure Of Enterococcus Faecium D63R Penicillin-Binding Protein 5 (Pbp5Fm)
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-04-24 Classification: ANTIBIOTIC Ligands: RB6, SO4 |
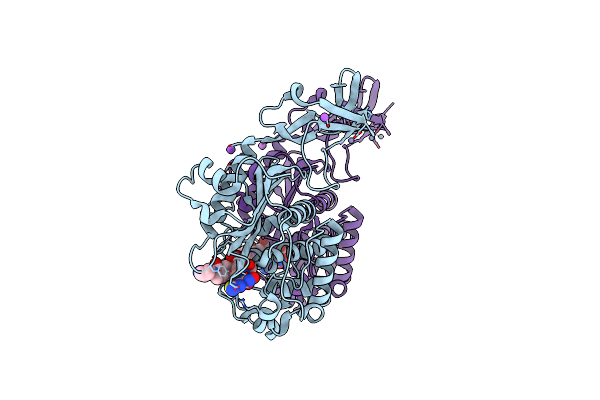 |
Crystal Structure Of S. Aureus Penicillin Binding Protein 4 (Pbp4) Mutant (E183A, F241R) In Complex With Ceftobiprole
Organism: Staphylococcus aureus (strain col)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-05-16 Classification: HYDROLASE / Antibiotic Ligands: ZN, RB6, NA |
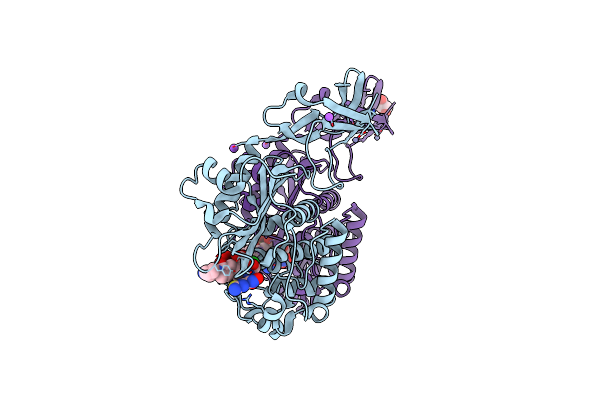 |
Crystal Structure Of Wild-Type S. Aureus Penicillin Binding Protein 4 (Pbp4) In Complex With Ceftobiprole
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-05-16 Classification: HYDROLASE / Antibiotic Ligands: RB6, SO4, ZN, CL, NA, GOL |
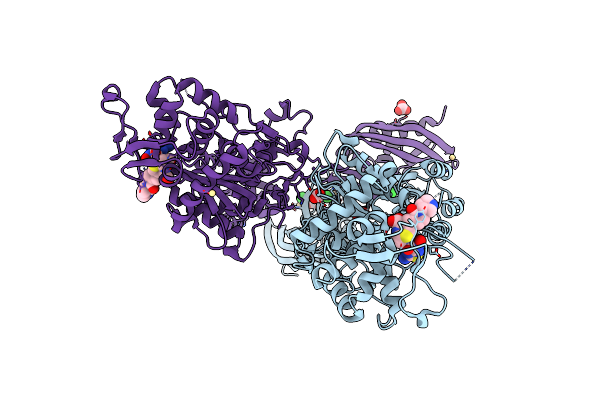 |
Structural Insights Into The Anti- Methicillin-Resistant Staphylococcus Aureus (Mrsa) Activity Of Ceftobiprole
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-08-01 Classification: HYDROLASE/Antibiotic Ligands: RB6, BCT, CD, CL |

